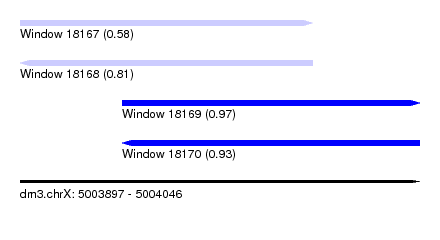
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,003,897 – 5,004,046 |
| Length | 149 |
| Max. P | 0.970412 |

| Location | 5,003,897 – 5,004,006 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Shannon entropy | 0.11149 |
| G+C content | 0.46789 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.30 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5003897 109 + 22422827 AUGUGCCUAAGUAGUUGCUGCUCUGGUAAUCAUGUUCGACACGCCCCCAUUUCGGGGGCAUUGCCAACAUGCCGCACCAACGAUUGUUGUUAUUAUUGGUGUUUAUAUU ...((((..(((((...)))))..))))..((((((.(....((((((.....))))))....).))))))..(((((((..((.......))..)))))))....... ( -37.00, z-score = -2.54, R) >droSim1.chrX_random 1748465 109 + 5698898 AUGUGCCUAAGUAGUUGCUGCUCUGGUAAUCGUGUUCGACACGCCCCCUUUUCGGGGGCGUUGCCAACGUGCCGCACCUGCGAUUGUUGUUAUUAUUGCUGUUUAUAUU ..........((((.(((.((.((((((((.(((.....)))((((((.....)))))))))))))..).)).))).))))............................ ( -35.40, z-score = -2.10, R) >droSec1.super_4 4426522 109 - 6179234 AUGUGCCUAAGUAGUUGCUGCUCUGGUAAUCGUGUUCGACACGCCCCCAUUUCGGGGGCGUUGCCAACAUGCCGCACCUACGAUUGUUGUUAUUAUUGCUGUUUAUAUU ..........((((.(((.((..(((((((.(((.....)))((((((.....)))))))))))))....)).))).))))............................ ( -35.50, z-score = -2.34, R) >droYak2.chrX 18304749 109 - 21770863 AUGUGCCUAAGUAGUUGCUGCUCUGGUAAUCAUGUUCGACACGCCCCCAUUUUGGGGGCGUUGGCUACAUGCAGCACCUUCGAUUGUUGUUAUUAUUGCUGUUUAUAUU ...((((..(((((...)))))..))))..(((((.....((((((((.....)))))))).....)))))(((((.....(((....))).....)))))........ ( -35.30, z-score = -2.28, R) >droEre2.scaffold_4690 2356129 109 + 18748788 AUGUGCCUAAGUAGUUGCUGCUCUGGUAAUCAUGUACGACACGCCCCCAUUUUGUGGGCGGUGGCUACAUGCCGCACCAUCGAUUGUUGUUAUUAUUGCUGUUUAUAUU ...((((..(((((...)))))..)))).......(((((((((((.(.....).)))))(((((.....))))).........))))))................... ( -31.80, z-score = -0.86, R) >consensus AUGUGCCUAAGUAGUUGCUGCUCUGGUAAUCAUGUUCGACACGCCCCCAUUUCGGGGGCGUUGCCAACAUGCCGCACCUACGAUUGUUGUUAUUAUUGCUGUUUAUAUU ...((((..(((((...)))))..))))..((((((.(.(((((((((.....))))))).)).)))))))...................................... (-27.34 = -27.30 + -0.04)

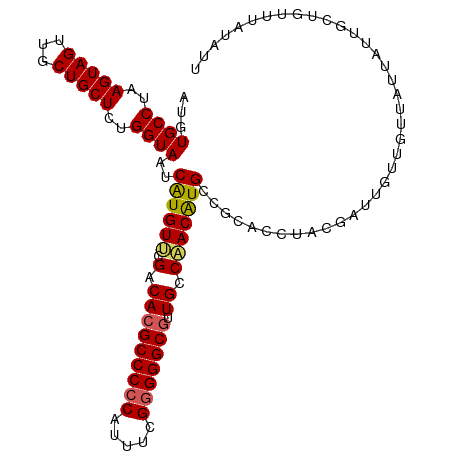
| Location | 5,003,897 – 5,004,006 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Shannon entropy | 0.11149 |
| G+C content | 0.46789 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5003897 109 - 22422827 AAUAUAAACACCAAUAAUAACAACAAUCGUUGGUGCGGCAUGUUGGCAAUGCCCCCGAAAUGGGGGCGUGUCGAACAUGAUUACCAGAGCAGCAACUACUUAGGCACAU ........((((((..((.......))..))))))...((((((((((.(((((((.....))))))))))).))))))....((.(((.((...)).))).))..... ( -34.30, z-score = -2.30, R) >droSim1.chrX_random 1748465 109 - 5698898 AAUAUAAACAGCAAUAAUAACAACAAUCGCAGGUGCGGCACGUUGGCAACGCCCCCGAAAAGGGGGCGUGUCGAACACGAUUACCAGAGCAGCAACUACUUAGGCACAU ........................(((((...((.((((((...(....)((((((.....)))))))))))).)).))))).((.(((.((...)).))).))..... ( -32.90, z-score = -2.72, R) >droSec1.super_4 4426522 109 + 6179234 AAUAUAAACAGCAAUAAUAACAACAAUCGUAGGUGCGGCAUGUUGGCAACGCCCCCGAAAUGGGGGCGUGUCGAACACGAUUACCAGAGCAGCAACUACUUAGGCACAU ............................((((.(((.((...((((..((((((((.....))))))))((((....))))..)))).)).))).)))).......... ( -33.20, z-score = -2.48, R) >droYak2.chrX 18304749 109 + 21770863 AAUAUAAACAGCAAUAAUAACAACAAUCGAAGGUGCUGCAUGUAGCCAACGCCCCCAAAAUGGGGGCGUGUCGAACAUGAUUACCAGAGCAGCAACUACUUAGGCACAU ..........((...............(....)((((((((((.....((((((((.....)))))))).....))))(.....)...)))))).........)).... ( -30.60, z-score = -2.37, R) >droEre2.scaffold_4690 2356129 109 - 18748788 AAUAUAAACAGCAAUAAUAACAACAAUCGAUGGUGCGGCAUGUAGCCACCGCCCACAAAAUGGGGGCGUGUCGUACAUGAUUACCAGAGCAGCAACUACUUAGGCACAU ..........((...............(..(((((...((((((.(.(((((((.(.....).))))).)).)))))))..)))))..)..............)).... ( -25.15, z-score = -0.04, R) >consensus AAUAUAAACAGCAAUAAUAACAACAAUCGAAGGUGCGGCAUGUUGGCAACGCCCCCGAAAUGGGGGCGUGUCGAACAUGAUUACCAGAGCAGCAACUACUUAGGCACAU ................................((((..((((((((((.(((((((.....))))))))))).)))))).......(((.((...)).)))..)))).. (-26.60 = -27.60 + 1.00)

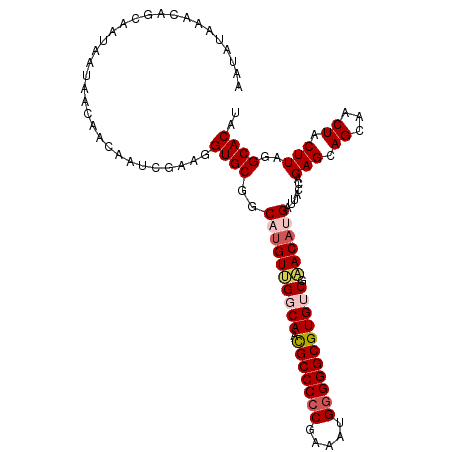
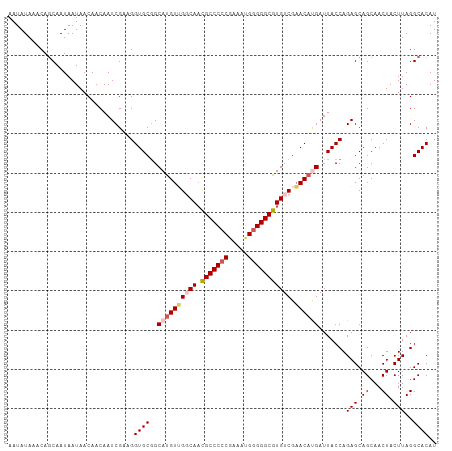
| Location | 5,003,935 – 5,004,046 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.90 |
| Shannon entropy | 0.16085 |
| G+C content | 0.44486 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -24.64 |
| Energy contribution | -26.32 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5003935 111 + 22422827 ACACGCCCCCAUUUCGGGGGCAUUGCCAACAUGCCGCACCAACGAUUGUUGUUAUUAUUGGUGUUUAUAUUGUUGCCGAAUUGUUAAUUGAUCUAAUUGAUAAAUUACCUG ....((((((.....)))))).(((.(((((....(((((((..((.......))..)))))))......))))).))).((((((((((...))))))))))........ ( -33.80, z-score = -3.11, R) >droSim1.chrX_random 1748503 111 + 5698898 ACACGCCCCCUUUUCGGGGGCGUUGCCAACGUGCCGCACCUGCGAUUGUUGUUAUUAUUGCUGUUUAUAUUGUUCCCGGCUUGGUAAUUGAUCUAAUUGAUGAAUUACCAG ..((((((((.....))))))))...(((((...(((....)))..)))))........((((.............)))).((((((((.(((.....))).)))))))). ( -39.02, z-score = -3.98, R) >droSec1.super_4 4426560 111 - 6179234 ACACGCCCCCAUUUCGGGGGCGUUGCCAACAUGCCGCACCUACGAUUGUUGUUAUUAUUGCUGUUUAUAUUGUUCCCGGCUUGGUAAUUGAUCUAAUUGAUGAAUUACCAG ..((((((((.....)))))))).........((((.....(((((....((.......)).......)))))...)))).((((((((.(((.....))).)))))))). ( -36.60, z-score = -3.39, R) >droYak2.chrX 18304787 111 - 21770863 ACACGCCCCCAUUUUGGGGGCGUUGGCUACAUGCAGCACCUUCGAUUGUUGUUAUUAUUGCUGUUUAUAUUGUUCCCGGCUUGGUAAUUGAUCUAAUUGAUGAACUGCCAG ....((((((.....))))))(((((..(((.((((((.....(((....))).....))))))......)))..))))).(((((.((.(((.....))).)).))))). ( -37.70, z-score = -3.25, R) >droEre2.scaffold_4690 2356167 109 + 18748788 ACACGCCCCCAUUUUGUGGGCGGUGGCUACAUGCCGCACCAUCGAUUGUUGUUAUUAUUGCUGUUUAUAUUGUUCCCGGCUUGGUAAUUGAUCUAAUUGAUGAACUACC-- ...(((((.(.....).)))))(((((.....)))))..(((((((((..(((((((..((((.............)))).))))))).....))))))))).......-- ( -32.52, z-score = -2.18, R) >consensus ACACGCCCCCAUUUCGGGGGCGUUGCCAACAUGCCGCACCUACGAUUGUUGUUAUUAUUGCUGUUUAUAUUGUUCCCGGCUUGGUAAUUGAUCUAAUUGAUGAAUUACCAG .(((((((((.....))))))).)).(((((...((......))..)))))........((((.............))))..(((((((.(((.....))).))))))).. (-24.64 = -26.32 + 1.68)

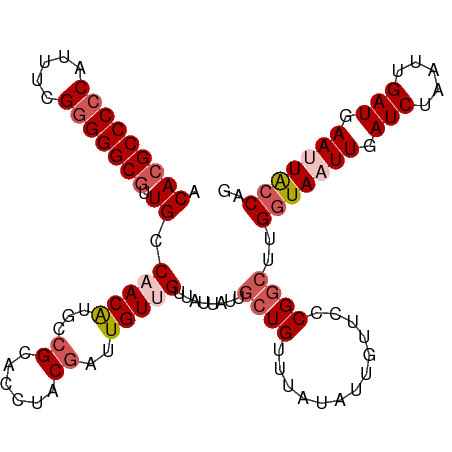
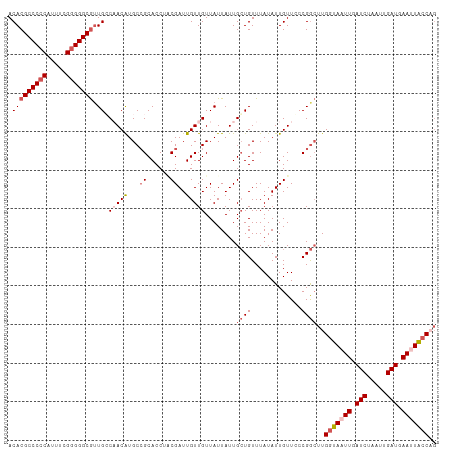
| Location | 5,003,935 – 5,004,046 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.90 |
| Shannon entropy | 0.16085 |
| G+C content | 0.44486 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -24.89 |
| Energy contribution | -25.73 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5003935 111 - 22422827 CAGGUAAUUUAUCAAUUAGAUCAAUUAACAAUUCGGCAACAAUAUAAACACCAAUAAUAACAACAAUCGUUGGUGCGGCAUGUUGGCAAUGCCCCCGAAAUGGGGGCGUGU ..(.(((((.(((.....))).))))).).....(.(((((.......((((((..((.......))..)))))).....))))).).((((((((.....)))))))).. ( -31.50, z-score = -2.22, R) >droSim1.chrX_random 1748503 111 - 5698898 CUGGUAAUUCAUCAAUUAGAUCAAUUACCAAGCCGGGAACAAUAUAAACAGCAAUAAUAACAACAAUCGCAGGUGCGGCACGUUGGCAACGCCCCCGAAAAGGGGGCGUGU .((((((((.(((.....))).)))))))).((((....).....................(((..((((....))))...)))))).((((((((.....)))))))).. ( -38.30, z-score = -4.09, R) >droSec1.super_4 4426560 111 + 6179234 CUGGUAAUUCAUCAAUUAGAUCAAUUACCAAGCCGGGAACAAUAUAAACAGCAAUAAUAACAACAAUCGUAGGUGCGGCAUGUUGGCAACGCCCCCGAAAUGGGGGCGUGU .((((((((.(((.....))).)))))))).((((....).....................((((.((((....))))..))))))).((((((((.....)))))))).. ( -36.50, z-score = -3.06, R) >droYak2.chrX 18304787 111 + 21770863 CUGGCAGUUCAUCAAUUAGAUCAAUUACCAAGCCGGGAACAAUAUAAACAGCAAUAAUAACAACAAUCGAAGGUGCUGCAUGUAGCCAACGCCCCCAAAAUGGGGGCGUGU .((((.(((((((.....)))......((.....))))))..((((..(((((....................)))))..))))))))((((((((.....)))))))).. ( -34.35, z-score = -2.94, R) >droEre2.scaffold_4690 2356167 109 - 18748788 --GGUAGUUCAUCAAUUAGAUCAAUUACCAAGCCGGGAACAAUAUAAACAGCAAUAAUAACAACAAUCGAUGGUGCGGCAUGUAGCCACCGCCCACAAAAUGGGGGCGUGU --(((((((.(((.....))).)))))))..(((((..............(((...((...........))..)))(((.....))).)).((((.....))))))).... ( -27.60, z-score = -1.04, R) >consensus CUGGUAAUUCAUCAAUUAGAUCAAUUACCAAGCCGGGAACAAUAUAAACAGCAAUAAUAACAACAAUCGAAGGUGCGGCAUGUUGGCAACGCCCCCGAAAUGGGGGCGUGU ..(((((((.(((.....))).)))))))..((((....)..........(((....................)))))).........((((((((.....)))))))).. (-24.89 = -25.73 + 0.84)

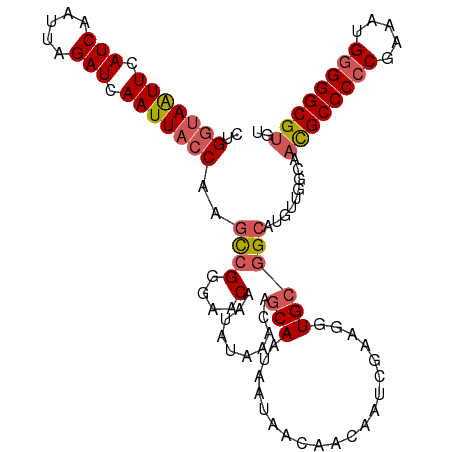
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:51 2011