
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,971,549 – 4,971,669 |
| Length | 120 |
| Max. P | 0.619639 |

| Location | 4,971,549 – 4,971,669 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.96 |
| Shannon entropy | 0.21609 |
| G+C content | 0.50160 |
| Mean single sequence MFE | -39.81 |
| Consensus MFE | -34.06 |
| Energy contribution | -32.95 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
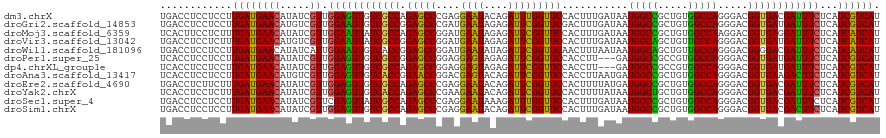
>dm3.chrX 4971549 120 + 22422827 UGACCUCCUCCUUGAUGAACAUAUCGUUGGAGUUGUCGCCAGAGCCCGAGGAAUACAGAUUUGGUUCCACUUUGAUAAUGGCCGCUGUGGCCAGGGACGGUGACGAUUUCUCAUCGUCAU ((((.........(((((.....)))))((((((((((((....(((..(((((.........)))))..........(((((.....))))))))..)))))))))))).....)))). ( -39.70, z-score = -1.22, R) >droGri2.scaffold_14853 4176320 120 + 10151454 UGACCUCCUCCUUGAUGAACAUGUCGUUGGAAUUGUCGCCGGAGCCCGAUGAAUAGAGAUUCGGUUCGACUUUGAUAAUGGCCGCUGUGGCCAGGGACGGUGAUGAUUUCUCAUCGUCAU ((((.........(((((.....)))))(((((..((((((...(((..(((((.(.....).)))))..........(((((.....)))))))).))))))..))))).....)))). ( -39.10, z-score = -0.78, R) >droMoj3.scaffold_6359 1200301 120 + 4525533 UCACUUCCUCUUUGAUGAACAUAUCGUUGGAAUUAUCGCCAGAGCCGGAUGAAUAGAGAUUCGGUUCCACUUUGAUAAUGGCCGCUGUGGCCAAGGACGGUGAGGAUUUCUCAUCAUCAU .....(((((((..((((.....))))..))...((((((.(((((((((........)))))))))...........(((((.....))))).)).))))))))).............. ( -36.90, z-score = -1.25, R) >droVir3.scaffold_13042 4936823 120 - 5191987 UGACCUCCUCUUUGAUGAACAUGUCGUUGGAAUUAUCGCCGGAGCCGGAUGAAUAGAGAUUCGGUUCCACUUUGAUAAUGGCAGCUGUUGCCAGGGACGGUGAUGAUUUCUCAUCAUCAU ............((((((...((.....((((((((((((((((((((((........))))))))))..........((((((...)))))).....)))))))))))).)))))))). ( -42.30, z-score = -2.42, R) >droWil1.scaffold_181096 4756602 120 + 12416693 UGACCUCCUCCUUGAUGAACAUAUCAUUGGAAUUGUCACCGGAGCCGGAUGAAUAUAGAUUCGGUUCAACUUUAAUAAUGGCAGCUGUUGCCAGGGACGGGGACGAUUUCUCAUCAUCAU ............((((((..........(((((((((.((((((((((((........)))))))))...........((((((...))))))....))).))))))))))))))).... ( -37.40, z-score = -1.72, R) >droPer1.super_25 76536 117 + 1448063 UCACCUCCUCCUUGAUGAACAUAUCGUUGGAGUUGUCGCCGGAGCCGGAGGAGUAGAGAUUCGGUUCCACCUU---GAUGGCCGCCGUGGCCAGGGACGGUGAUGAUUUCUCAUCGUCAU ............((((((((.....)).(((((..((((((((((((((..........))))))))).(((.---..(((((.....))))))))..)))))..)))))...)))))). ( -43.80, z-score = -1.06, R) >dp4.chrXL_group1e 6740746 117 + 12523060 UCACCUCCUCCUUGAUGAACAUAUCGUUGGAGUUGUCGCCAGAGCCGGAGGAGUAGAGAUUCGGUUCCACCUU---GAUGGCCGCCGUGGCCAGGGACGGUGAUGAUUUCUCAUCGUCAU ((..(((((((..(((((.....)))))((..(((....)))..)))))))))..)).....((.....))..---..(((((.....)))))..((((((((.(....))))))))).. ( -39.90, z-score = -0.35, R) >droAna3.scaffold_13417 335394 120 + 6960332 UCACCUCCUCCUUGAUGAACAUGUCGUUGGAGUUGUCACCGGAACCGGACGAGUACAGAUUCGGUUCCACCUUAAUGAUGGCCGCUGUGGCCAGGGACGGUGAAGACUUCUCAUCGUCAU (((((..((((.((((......))))..))))..(((.((((((((((((.......).)))))))))..........(((((.....))))))))))))))).(((........))).. ( -45.30, z-score = -1.97, R) >droEre2.scaffold_4690 2331154 120 + 18748788 UGACCUCUUCUUUGAUGAACAUAUCGUUGGAGUUGUCGCCAGAGCCCGAGGAAUACAGAUUCGGUUCCACUUUUAUGAUGGCCGCUGUGGCCAGGGACGGUGACGAUUUCUCAUCGUCAU ((((.........(((((.....)))))((((((((((((....(((..(((((.........)))))..........(((((.....))))))))..)))))))))))).....)))). ( -39.70, z-score = -1.27, R) >droYak2.chrX 18279934 120 - 21770863 UCACCUCCUCUUUGAUGAACAUAUCGUUGGAGUUGUCACCAGAGCCCGAAGAAUACAGAUUCGGUUCCACUUUUAUAAUGGCUGCUGUGGCCAGGGACGGUGACGAUUUCUCAUCGUCAU ..(((....(((..((((.....))))..)))..(((.((.(((((.(((.........))))))))...........(((((.....)))))))))))))((((((.....)))))).. ( -35.90, z-score = -1.18, R) >droSec1.super_4 4403616 120 - 6179234 UGACCUCCUCCUUGAUGAACAUAUCGUUCGAGUUAUCGCCAGAGCCCGAGGAAUAAAGAUUUGGUUCCACUUUGAUAAUGGCCGCUGUGGCCAGGGACGGUGACGAUUUCUCAUCGUCAU ((((............((((.....))))(((..((((.((..((((..(((((.........)))))..........(((((.....)))))))).)..)).))))..)))...)))). ( -35.70, z-score = -0.74, R) >droSim1.chrX 3848972 120 + 17042790 UGACCUCCUCCUUGAUGAACAUAUCGUUGGAGUUGUCGCCAGAGCCCGAGGAAUACAGAUUCGGUUCCACUUUGAUAAUGGCCGCUGUGGCCAGGGACGGUGACGACUUCUCAUCGUCAU ((((.........(((((.....)))))((((((((((((....(((..(((((.........)))))..........(((((.....))))))))..)))))))))))).....)))). ( -42.00, z-score = -1.59, R) >consensus UGACCUCCUCCUUGAUGAACAUAUCGUUGGAGUUGUCGCCAGAGCCCGAGGAAUACAGAUUCGGUUCCACUUUGAUAAUGGCCGCUGUGGCCAGGGACGGUGACGAUUUCUCAUCGUCAU ............((((((((.....)).((((((((((((.(((((....((((....)))))))))...........(((((.....))))).....))))))))))))...)))))). (-34.06 = -32.95 + -1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:46 2011