
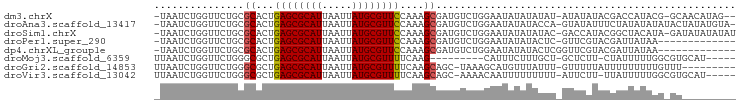
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,881,084 – 4,881,176 |
| Length | 92 |
| Max. P | 0.963810 |

| Location | 4,881,084 – 4,881,176 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 67.66 |
| Shannon entropy | 0.63692 |
| G+C content | 0.37755 |
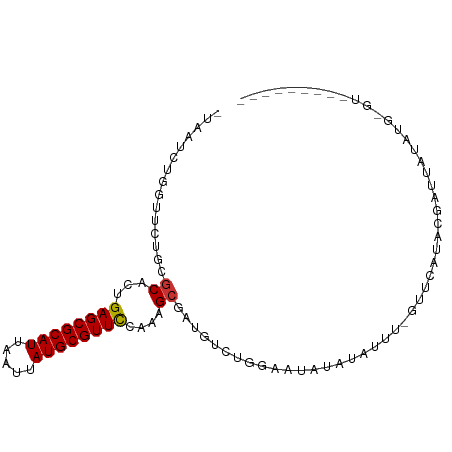
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963810 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 4881084 92 - 22422827 -UAAUCUGGUUCUGCGCACUGAGCGCAUUAAUUAUGCGUUCCAAAGCGAUGUCUGGAAUAUAUAUAU-AUAUAUACGACCAUACG-GCAACAUAG-- -...((..(.....(((...((((((((.....))))))))....)))....)..))((((((....-))))))..........(-....)....-- ( -21.50, z-score = -1.01, R) >droAna3.scaffold_13417 237982 94 - 6960332 -UAAUCUGGUUCUGCGCACUGAGCGCAUUAAUUAUGCGUUCCAAAGCGAUGUCUGGAAUAUAUACCA-GUAUAUUUCUAUAUAUAUACUAUAUGUA- -...((..(.....(((...((((((((.....))))))))....)))....)..)).((((((..(-((((((........))))))).))))))- ( -25.90, z-score = -2.63, R) >droSim1.chrX 3782190 94 - 17042790 -UAAUCUGGUUCUGCGCACUGAGCGCAUUAAUUAUGCGUUCCAAAGCGAUGUCUGGAAUAUAUAUAC-GACCAUACGGCUACAUA-GAUAUAUAUAU -...((..(.....(((...((((((((.....))))))))....)))....)..))((((((((..-..((....)).......-.)))))))).. ( -22.60, z-score = -1.04, R) >droPer1.super_290 17986 82 - 47041 -UAAUCUGGUUCUGCGCACUGAGCGCAUUAAUUAUGCGUUCCAAAGCGAUGUCUGGAAUAUAUACUC-GUUCGUACGAUUAUAA------------- -...((..(.....(((...((((((((.....))))))))....)))....)..))...((((.((-((....)))).)))).------------- ( -23.70, z-score = -2.38, R) >dp4.chrXL_group1e 6666017 83 - 12523060 -UAAUCUGGUUCUGCGCACUGAGCGCAUUAAUUAUGCGUUCCAAAGCGAUGUCUGGAAUAUAUACUCGGUUCGUACGAUUAUAA------------- -...((..(.....(((...((((((((.....))))))))....)))....)..))...((((.(((.......))).)))).------------- ( -22.80, z-score = -1.79, R) >droMoj3.scaffold_6359 1062077 81 - 4525533 UUAAUCUGGUUCUGGGCGCUGAGCGCAUUAAUUAUGCGUUUUCAAG---------CAUUUCUUUGCU-GCUCUU-CUAUUUUUGGCGUGCAU----- ............((.(((((((((((((.....))))))))...((---------((......))))-......-........))))).)).----- ( -21.70, z-score = -1.35, R) >droGri2.scaffold_14853 4076124 86 - 10151454 UUAAUCUGGUUCUGGGCGCUGAGCGCAUUAAUUAUGCGUUUUCAAGCAGC-UAAAGCAUGUUUAUUU-GUUUUUAUUUUUUUUUGUUU--------- ..............((((((((((((((.....))))))))...))).))-)((((((........)-)))))...............--------- ( -16.90, z-score = -0.42, R) >droVir3.scaffold_13042 4797482 89 + 5191987 UUAAUCUGGUUCUGGGCGCUGAGCGCAUUAAUUAUGCGUUUUCAAGCAGC-AAAACAAUUUUUUUUU-AUUCUU-UUAUUUUUGGCGUGCAU----- ............((.(((((((((((((.....)))))))).........-((((.(((........-))).))-))......))))).)).----- ( -16.60, z-score = 0.23, R) >consensus _UAAUCUGGUUCUGCGCACUGAGCGCAUUAAUUAUGCGUUCCAAAGCGAUGUCUGGAAUAUAUAUUU_GUUCAUACGAUUAUAUG_GU_________ ...............((...((((((((.....))))))))....)).................................................. (-11.70 = -11.59 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:40 2011