
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,868,790 – 4,868,883 |
| Length | 93 |
| Max. P | 0.622763 |

| Location | 4,868,790 – 4,868,883 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 97 |
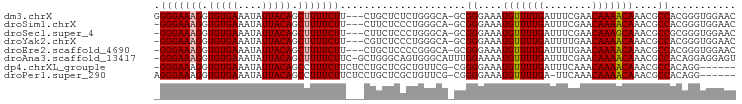
| Reading direction | reverse |
| Mean pairwise identity | 85.94 |
| Shannon entropy | 0.27851 |
| G+C content | 0.45644 |
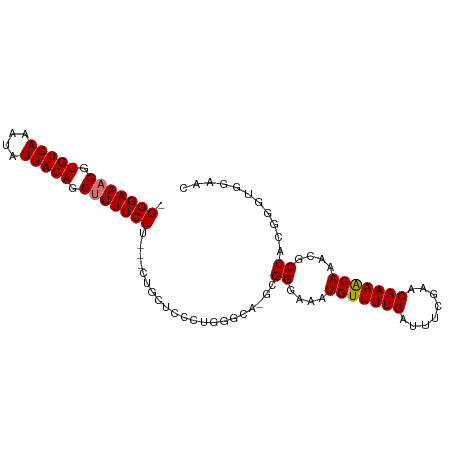
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -16.82 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4868790 93 - 22422827 GGGGAAAGGUGUGAAAUAUUACAGCUUUUCUU---CUGCUCUCUGGGCA-GCGGGAAAUGUUUUGAUUUCGAACAAAACAAACGCCACGGGUGGAAC ((((((((.(((((....))))).))))))))---(((((.....))))-)((((...(((((((........)))))))....)).))........ ( -26.50, z-score = -1.99, R) >droSim1.chrX 3769153 92 - 17042790 -GGGAAAGGUGUGAAAUAUUACAGCUUUUCUU---CUUCUCCCUGGGCA-GCGGGAAAUGUUUUGAUUUCGAACAAAACAAACGCCACGGGUGGAAC -(((((((.(((((....))))).))))))).---.(((..((((....-(((.....(((((((........)))))))..)))..))))..))). ( -27.70, z-score = -2.04, R) >droSec1.super_4 4309203 92 + 6179234 -GGGAAAGGUGUGAAAUAUUACAGCUUUUCUU---CUUCUCCCUGGGCA-GCGGGAAAUGUUUUGAUUUCGAACAAAGCAAACGCCGCGGGUGGAAC -(((((((.(((((....))))).))))))).---.(((..((((.(..-(((.....(((((((........)))))))..)))).))))..))). ( -28.50, z-score = -1.65, R) >droYak2.chrX 18184326 92 + 21770863 -GGGAAAGGUGUGAAAUAUUACAGCUUUUCUU---CGUCUCCCUGGGCA-GCGGGAAAUGUUUUGAUUUUGAACAAAACAAACGCCACGGGUGGAAC -(((((((.(((((....))))).))))))).---..((..((((....-(((.....(((((((........)))))))..)))..))))..)).. ( -27.20, z-score = -1.79, R) >droEre2.scaffold_4690 2234124 92 - 18748788 -GGGAAAGGUGUGAAAUAUUACAGCUUUUCUU---CUGCUCCCCGGGCA-GCGGGAAAUGUUUUGAUUUUGAACAAAACAAACGCCACGGGUGGAAC -(((((((.(((((....))))).)))))))(---(..(((((((....-.))))...(((((((........)))))))........)))..)).. ( -29.50, z-score = -2.27, R) >droAna3.scaffold_13417 224570 95 - 6960332 -GGGAAAGGUGUGAAAUAUUACAGCUUUUCUUC-GCUGGGCAGUGGGCAUUUGGAAAAUGUUUUGAUUUCGAACAAAACAAACGCCACAGGAGGAGU -...((((.(((((....))))).))))(((((-.((((((.(....)..........(((((((........)))))))...))).)))))))).. ( -25.20, z-score = -1.63, R) >dp4.chrXL_group1e 6650181 89 - 12523060 -GGGAAAGGUGUGAAAUAUUACAGCCUUUCUUCUCCUGCUCGCUGUUCG-CGGGGAAAUGUUUUGAUUUCAAACAAAACAAACGCCACAGG------ -(((((((((((((....))))).))))))))..(((((((((.....)-))))....(((((((........))))))).......))))------ ( -27.30, z-score = -2.67, R) >droPer1.super_290 2252 89 - 47041 AGGGAAAGGUGUGAAAUAUUACAGCCUUUCUUCUCCUGCUCGCUGUUCG-CGGGGAAAUGUUUUGA-UUCAAACAAAACAAACGCCACAGG------ .(((((((((((((....))))).))))))))..(((((((((.....)-))))....(((((((.-......))))))).......))))------ ( -27.30, z-score = -2.50, R) >consensus _GGGAAAGGUGUGAAAUAUUACAGCUUUUCUU___CUGCUCCCUGGGCA_GCGGGAAAUGUUUUGAUUUCGAACAAAACAAACGCCACGGGUGGAAC .(((((((.(((((....))))).))))))).....................((....(((((((........)))))))....))........... (-16.82 = -16.96 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:37 2011