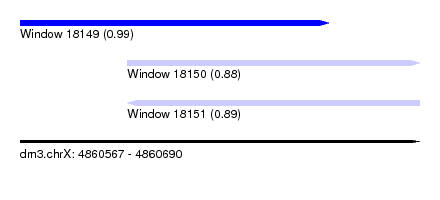
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,860,567 – 4,860,690 |
| Length | 123 |
| Max. P | 0.994202 |

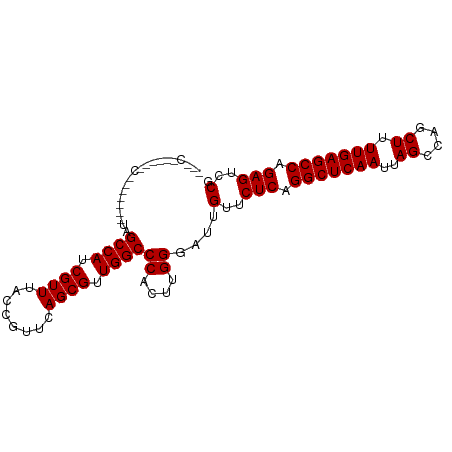
| Location | 4,860,567 – 4,860,662 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Shannon entropy | 0.23168 |
| G+C content | 0.53627 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -26.38 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
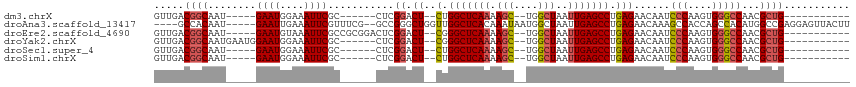
>dm3.chrX 4860567 95 + 22422827 ---CGACUUGCACAGCCUUAGCCAUCGUUUACCGUUCAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCAGCUUUUGAGCCAGAGUCCG ---.((((((.(((((((..((((.((((........)))).))))......))).)))).)...(((((((..((....)).))))))).))))).. ( -29.90, z-score = -2.09, R) >droEre2.scaffold_4690 2225995 92 + 18748788 CGACUUGCAACC------UAGCCAUCGUUUACCGUUCAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCAGCUUUUGAGCCCGAGUCCG .((((((.....------..((((.((((........)))).)))).....(((((....)))))(((((((..((....)).))))))))))))).. ( -30.50, z-score = -2.74, R) >droYak2.chrX 18175938 98 - 21770863 CGACUUGCAACCCAGCCAUAGCCAUCGUUUACCGUUCAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCAGCUUUUGAGCCCGAGUCCG .((((((((((((((.....((((.((((........)))).))))....))))).)))......(((((((..((....)).))))))))))))).. ( -33.70, z-score = -3.48, R) >droSec1.super_4 4301531 80 - 6179234 ------------------UAGCCAUCGUUUACCGUUCAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCAGCUUUUGAGCCAGAGUCCG ------------------..((((.((((........)))).))))((....))....(..(((.(((((((..((....)).))))))).)))..). ( -26.10, z-score = -2.63, R) >droSim1.chrX 3761177 80 + 17042790 ------------------UAGCCAUCGUUUACCGUUCAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCAGCUUUUGAGCCAGAGUCCG ------------------..((((.((((........)))).))))((....))....(..(((.(((((((..((....)).))))))).)))..). ( -26.10, z-score = -2.63, R) >consensus ___C______C_______UAGCCAUCGUUUACCGUUCAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCAGCUUUUGAGCCAGAGUCCG ....................((((.((((........)))).))))((....))....(..(((.(((((((..((....)).))))))).)))..). (-26.38 = -26.38 + 0.00)

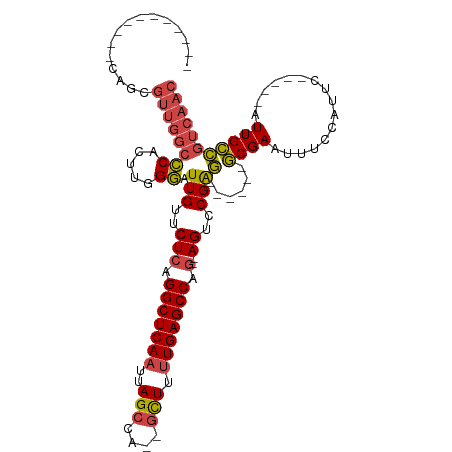
| Location | 4,860,600 – 4,860,690 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Shannon entropy | 0.32551 |
| G+C content | 0.52165 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4860600 90 + 22422827 -----------CAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCA--GCUUUUGAGCCAG--AGUCCGAG------GCGAAUUUCCAUUC-----AUUGCCGUCAAC -----------....((((((((....))..(((..(((.(((((((..((...--.)).))))))).)--))..)))(------((((..........-----.))))))))))) ( -29.50, z-score = -2.02, R) >droAna3.scaffold_13417 212790 105 + 6960332 AAGUAACUCCUCGGCCAUGUGGCUGGUGGCUUUGUUCUCAGGCUCAAUUAGCCAUUAUUUGUGAGCCAACCAGCCCGGC--CGAAACGAAUUUCAAUUC-----AUUGUGGC---- ........((((((((..(.((((((((((((........((((.....)))).........))))).)))))))))))--)))...((((....))))-----.....)).---- ( -37.83, z-score = -2.92, R) >droEre2.scaffold_4690 2226025 96 + 18748788 -----------CAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCA--GCUUUUGAGCCCG--AGUCCGAGUCCGCGGCGAAUUUACAUUC-----AUUGCCGUCAAC -----------....((((((......((..(((..(((.(((((((..((...--.)).))))))).)--))..)))..))..(((((..........-----.))))))))))) ( -34.10, z-score = -2.67, R) >droYak2.chrX 18175974 95 - 21770863 -----------CAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCA--GCUUUUGAGCCCG--AGUCCGAG------GCGAAUUUCCAUUCCAUUCAUUGCCGUCAAC -----------....((((((((....))..(((..(((.(((((((..((...--.)).))))))).)--))..)))(------((((................))))))))))) ( -29.79, z-score = -1.97, R) >droSec1.super_4 4301549 90 - 6179234 -----------CAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCA--GCUUUUGAGCCAG--AGUCCGAG------GCGAAUUUCCAUUC-----AUUGCCGUCAAC -----------....((((((((....))..(((..(((.(((((((..((...--.)).))))))).)--))..)))(------((((..........-----.))))))))))) ( -29.50, z-score = -2.02, R) >droSim1.chrX 3761195 90 + 17042790 -----------CAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCA--GCUUUUGAGCCAG--AGUCCGAG------GCGAAUUUCCAUUC-----AUUGCCGUCAAC -----------....((((((((....))..(((..(((.(((((((..((...--.)).))))))).)--))..)))(------((((..........-----.))))))))))) ( -29.50, z-score = -2.02, R) >consensus ___________CAGCGUUGGCCCACUUGGGAUUGUUCUCAGGCUCAAUUAGCCA__GCUUUUGAGCCAG__AGUCCGAG______GCGAAUUUCCAUUC_____AUUGCCGUCAAC ...............((((((...((((((..((....))(((((((..(((....))).)))))))......))))))......((((................)))).)))))) (-18.88 = -19.86 + 0.98)

| Location | 4,860,600 – 4,860,690 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Shannon entropy | 0.32551 |
| G+C content | 0.52165 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -19.57 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
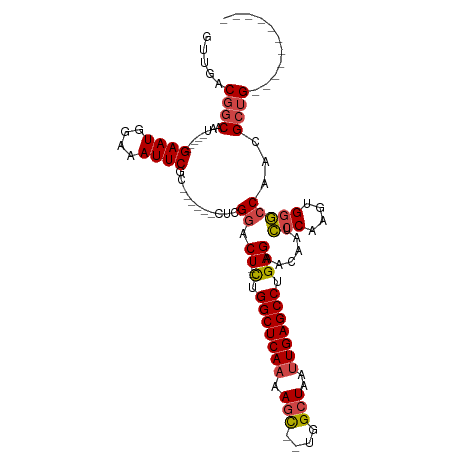
>dm3.chrX 4860600 90 - 22422827 GUUGACGGCAAU-----GAAUGGAAAUUCGC------CUCGGACU--CUGGCUCAAAAGC--UGGCUAAUUGAGCCUGAGAACAAUCCCAAGUGGGCCAACGCUG----------- ((((..(((...-----((((....))))))------)..(..((--(.(((((((.((.--...))..))))))).)))..)...(((....))).))))....----------- ( -29.10, z-score = -1.85, R) >droAna3.scaffold_13417 212790 105 - 6960332 ----GCCACAAU-----GAAUUGAAAUUCGUUUCG--GCCGGGCUGGUUGGCUCACAAAUAAUGGCUAAUUGAGCCUGAGAACAAAGCCACCAGCCACAUGGCCGAGGAGUUACUU ----.((..(((-----((((....)))))))(((--(((.(((((((.((((..........((((.....)))).........)))))))))))....))))))))........ ( -41.31, z-score = -3.99, R) >droEre2.scaffold_4690 2226025 96 - 18748788 GUUGACGGCAAU-----GAAUGUAAAUUCGCCGCGGACUCGGACU--CGGGCUCAAAAGC--UGGCUAAUUGAGCCUGAGAACAAUCCCAAGUGGGCCAACGCUG----------- (((..((((...-----((((....))))))))..)))..((.((--(((((((((.((.--...))..)))))))))))......(((....))))).......----------- ( -33.70, z-score = -2.14, R) >droYak2.chrX 18175974 95 + 21770863 GUUGACGGCAAUGAAUGGAAUGGAAAUUCGC------CUCGGACU--CGGGCUCAAAAGC--UGGCUAAUUGAGCCUGAGAACAAUCCCAAGUGGGCCAACGCUG----------- .....((((........((((....))))..------...((.((--(((((((((.((.--...))..)))))))))))......(((....)))))...))))----------- ( -32.30, z-score = -2.11, R) >droSec1.super_4 4301549 90 + 6179234 GUUGACGGCAAU-----GAAUGGAAAUUCGC------CUCGGACU--CUGGCUCAAAAGC--UGGCUAAUUGAGCCUGAGAACAAUCCCAAGUGGGCCAACGCUG----------- ((((..(((...-----((((....))))))------)..(..((--(.(((((((.((.--...))..))))))).)))..)...(((....))).))))....----------- ( -29.10, z-score = -1.85, R) >droSim1.chrX 3761195 90 - 17042790 GUUGACGGCAAU-----GAAUGGAAAUUCGC------CUCGGACU--CUGGCUCAAAAGC--UGGCUAAUUGAGCCUGAGAACAAUCCCAAGUGGGCCAACGCUG----------- ((((..(((...-----((((....))))))------)..(..((--(.(((((((.((.--...))..))))))).)))..)...(((....))).))))....----------- ( -29.10, z-score = -1.85, R) >consensus GUUGACGGCAAU_____GAAUGGAAAUUCGC______CUCGGACU__CUGGCUCAAAAGC__UGGCUAAUUGAGCCUGAGAACAAUCCCAAGUGGGCCAACGCUG___________ .....((((........((((....))))...........((.......(((((((.(((....)))..)))))))..........(((....)))))...))))........... (-19.57 = -20.15 + 0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:36 2011