
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,839,709 – 4,839,870 |
| Length | 161 |
| Max. P | 0.979550 |

| Location | 4,839,709 – 4,839,870 |
|---|---|
| Length | 161 |
| Sequences | 4 |
| Columns | 163 |
| Reading direction | forward |
| Mean pairwise identity | 82.93 |
| Shannon entropy | 0.27937 |
| G+C content | 0.38864 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -24.61 |
| Energy contribution | -27.55 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
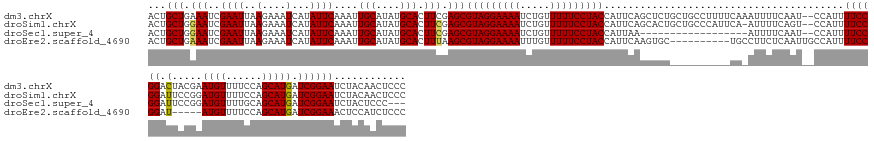
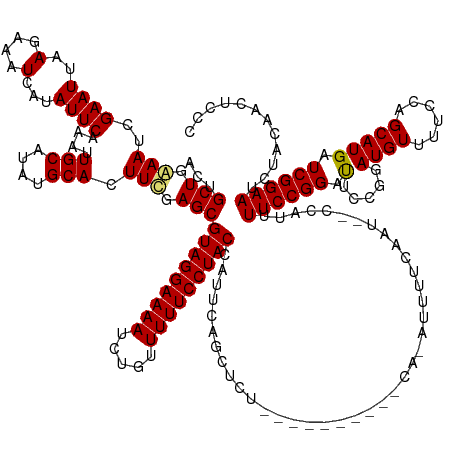
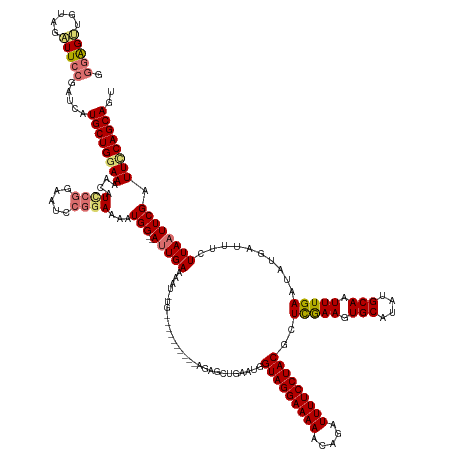
>dm3.chrX 4839709 161 + 22422827 ACUGCUGAAAUCGAAUUAAGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGCUCUGCUGCCUUUUCAAAUUUUCAAU--CCAUUUUCCGGACUACGAAUGUUUUCCAGCAUGAUCGGAAUCUACAACUCCC ...((((((.(((((....(((......)))....(((....))).)))))(.(((((((((.....)))))))))).))))))(((((((.................(--((.......)))....(((....)))))))).))..(((.........))). ( -34.40, z-score = -2.13, R) >droSim1.chrX 3742300 160 + 17042790 ACUGCUGGAAUCGAAUUAAGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGCACUGCUGCCCAUUCA-AUUUUCAGU--CCAUUUUCCGGAUUCCGGAUGUUUUCCAGCAUGAUCGGAAUCUACAACUCCC ..(((((((.(((((....(((......)))....(((....))).)))))(.(((((((((.....)))))))))).)))))))..((((........-.....))))--.........(((((((((((((......))))..)))))))))......... ( -39.72, z-score = -2.40, R) >droSec1.super_4 4281104 140 - 6179234 ACUGCUGGAAUCGAAUUAAGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAUUAA------------------AUUUUCAAU--CCAUUUUCCGGAUUCCGGAUGUUUUGCAGCAUGAUCGGAAUCUACUCCC--- .....((((.(((((....(((......)))....(((....))).)))))(.(((((((((.....)))))))))).....------------------........)--)))......(((((((((((((......))))..)))))))))......--- ( -34.90, z-score = -2.28, R) >droEre2.scaffold_4690 2202506 148 + 18748788 ACUGCUGAAAUCGAAUUAAGAAAUCAUAUUCAAAUUGCAUAUGCACUUUAAGCGUAGGAAAAUUUGUUUUUCCUACCAUUCAAGUGC----------UGCCUUCUCAAUUGCCAUUUUCCGGAU-----AUGUUUUCCAGCAUGAUCGGAAACUCCAUCUCCC ..(((((............((((.(((((((((((.(((...((((((.((..(((((((((.....)))))))))..)).))))))----------............))).))))...))))-----))).))))))))).(((.((.....))))).... ( -34.96, z-score = -2.85, R) >consensus ACUGCUGAAAUCGAAUUAAGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGCUCU__________CA_AUUUUCAAU__CCAUUUUCCGGAUUCCGGAUGUUUUCCAGCAUGAUCGGAAUCUACAACUCCC ...((((((.(((((....(((......)))....(((....))).)))))(.(((((((((.....)))))))))).))))))....................................(((((((((((((......))))..)))))))))......... (-24.61 = -27.55 + 2.94)
| Location | 4,839,709 – 4,839,870 |
|---|---|
| Length | 161 |
| Sequences | 4 |
| Columns | 163 |
| Reading direction | reverse |
| Mean pairwise identity | 82.93 |
| Shannon entropy | 0.27937 |
| G+C content | 0.38864 |
| Mean single sequence MFE | -44.35 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
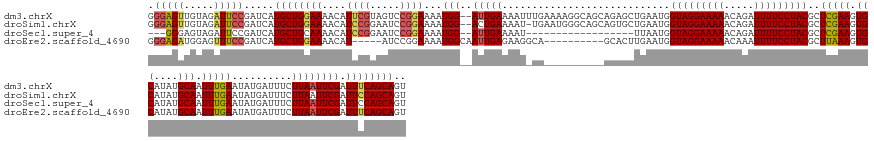
>dm3.chrX 4839709 161 - 22422827 GGGAGUUGUAGAUUCCGAUCAUGCUGGAAAACAUUCGUAGUCCGGAAAAUGG--AUUGAAAAUUUGAAAAGGCAGCAGAGCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCUUAAUUCGAUUUCAGCAGU .((((((...)))))).....((((((((..(((((.(((((((.....)))--))))............(((......))))))))(((((((((.....)))))))))..(((((.(((....)))....(((......)))....))))))))))))).. ( -44.60, z-score = -2.09, R) >droSim1.chrX 3742300 160 - 17042790 GGGAGUUGUAGAUUCCGAUCAUGCUGGAAAACAUCCGGAAUCCGGAAAAUGG--ACUGAAAAU-UGAAUGGGCAGCAGUGCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCUUAAUUCGAUUCCAGCAGU .((((((...)))))).....((((((((....(((((...)))))......--........(-((((((((((((...))).....(((((((((.....)))))))))))))(((((.(((((.(.....).))))))))))...)))))))))))))).. ( -50.00, z-score = -2.91, R) >droSec1.super_4 4281104 140 + 6179234 ---GGGAGUAGAUUCCGAUCAUGCUGCAAAACAUCCGGAAUCCGGAAAAUGG--AUUGAAAAU------------------UUAAUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCUUAAUUCGAUUCCAGCAGU ---((((.....))))......(((((......(((((...)))))...(((--((((((...------------------......(((((((((.....)))))))))..(((((.(((....))).)))))..............))))).))))))))) ( -41.20, z-score = -3.05, R) >droEre2.scaffold_4690 2202506 148 - 18748788 GGGAGAUGGAGUUUCCGAUCAUGCUGGAAAACAU-----AUCCGGAAAAUGGCAAUUGAGAAGGCA----------GCACUUGAAUGGUAGGAAAAACAAAUUUUCCUACGCUUAAAGUGCAUAUGCAAUUUGAAUAUGAUUUCUUAAUUCGAUUUCAGCAGU ..(((((.(((((...((((((.(((((......-----.))))).........(((.(((..(((----------((((((....((((((((((.....))))))))).)...))))))...)))..))).)))))))))....))))).)))))...... ( -41.60, z-score = -2.65, R) >consensus GGGAGUUGUAGAUUCCGAUCAUGCUGGAAAACAUCCGGAAUCCGGAAAAUGG__AUUGAAAAU_UG__________AGAGCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCUUAAUUCGAUUCCAGCAGU .(((((.....))))).....((((((((....((((.....))))...(((.....((((..........................(((((((((.....)))))))))..(((((.(((....))).)))))......)))).....))).)))))))).. (-31.48 = -31.98 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:33 2011