| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,125,387 – 10,125,477 |
| Length | 90 |
| Max. P | 0.542642 |

| Location | 10,125,387 – 10,125,477 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.74 |
| Shannon entropy | 0.54217 |
| G+C content | 0.64207 |
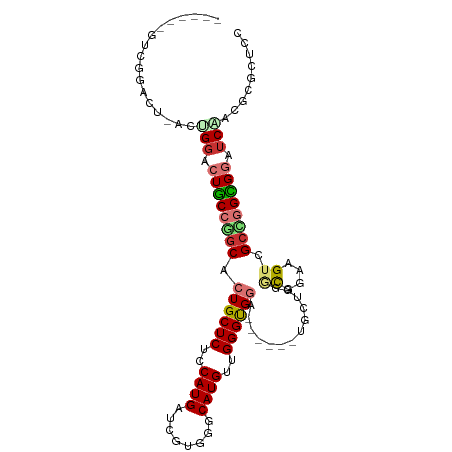
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.96 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10125387 90 - 23011544 ------UUCGGAGU---UGGACUGCCGGCACUGCUCUCCAUGAUCGUUGGCAUGUUGGGUGGA------UGCUGGGCGGAAGUCGCCGGCGGAUCAACGCGCUCC ------...(((((---(((.((((((((.(..(((..((((........))))..)))..).------.....(((....))))))))))).)))....))))) ( -41.00, z-score = -2.04, R) >droSim1.chr2L 9916570 99 - 22036055 UUCGGAGUGGGGCUAACUGGACUGCCGGCACUGCUCUCCAUGAUCGUGGGCAUGUUGGGUGGA------UGCUGUGCCGAAGUCGCCGGCGGAUCAACACGCUCC ...((((((((.....)).((((((((((.(..(((..((((........))))..)))..).------.(((.......))).)))))))).))....)))))) ( -38.30, z-score = -0.29, R) >droSec1.super_3 5563418 99 - 7220098 UUCGGAGUGGGGCUAACUGGACUGCCGGCACUGCUCUCCAUGAUCGUGGGCAUGUUGGGUGGA------UGCUGUGCAGAAGUCGCCGGCGGAUCAACACGCUCC ...((((((((.....)).((((((((((.(..(((..((((........))))..)))..).------(((...)))......)))))))).))....)))))) ( -38.80, z-score = -0.64, R) >droYak2.chr2L 12810268 99 - 22324452 UUGGGGGUCGGAGUGACUGGACUGCCUGCACUGCUCUCCAUGAUCGUGGGCAUGUUGGGUGGA------UGUGGGGCGGAAGUCGCCGGCGGAUCUACGCGCUCC .........((((((..(((((((((.((((..(((..((((........))))..)))..).------)))(((((....))).))))))).))))..)))))) ( -42.00, z-score = -1.03, R) >droEre2.scaffold_4929 10740426 99 - 26641161 UUCGGAGCCGGACUGACUGGACUGCCAGCGCUGCUCUCCAUGAUCGUGGGCAUGUUGGGCGGA------UGUUGGACGGAAGUCGCCGGCGGAUCCACGCGCUCC ...((((((((...((((...(((((((((((((((..((((........))))..)))))).------)))))).))).)))).)))(((......)))))))) ( -45.70, z-score = -1.75, R) >droAna3.scaffold_12916 13958777 99 - 16180835 ------CUGGGAGUCGGGGGACUUCCGGCACUGCUCUCCAUGAUCGUGGGCAUGUUGGGUGGAUGGGCGUGGUUGGUCGAUGUGGCCGGAGGAUCCACCCGCUCC ------...((((.(((((((((((((((.(..(((..((((........))))..)))..)....((((.(.....).)))).)))))))).))).)))))))) ( -47.90, z-score = -1.92, R) >droVir3.scaffold_12723 2002687 81 + 5802038 -------UUGGGC--GCCGGGCUGCCGGCACUGCUCUCCAUGAUGGUAGGCAUGUUGGGCGCA------UGCUGGGCCAGCGGCGCUUGUGUCUCC--------- -------..((((--(((((((..((((((.(((((..((((........))))..)))))..------)))))))))..))))))))........--------- ( -36.90, z-score = -0.09, R) >droGri2.scaffold_15252 3848413 90 - 17193109 -------UUGGGC--GAUGCGCUGCCAGCACUGCUCUCCAUGAUGGUGGGCAUGUUGGGCGUG------UGUUGAGCGAGCGACGGUUGCGCUUCGCUGCGAUCU -------...(((--((.((((.(((.((((.((((..((((........))))..)))))))------)((((......))))))).)))).)))))....... ( -37.40, z-score = -0.31, R) >consensus ______GUCGGACU_ACUGGACUGCCGGCACUGCUCUCCAUGAUCGUGGGCAUGUUGGGUGGA______UGCUGGGCGGAAGUCGCCGGCGGAUCAACGCGCUCC .................(((.((((((((.((((((..((((........))))..)))))).............((....)).)))))))).)))......... (-21.19 = -21.96 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:42 2011