
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,826,754 – 4,826,823 |
| Length | 69 |
| Max. P | 0.999488 |

| Location | 4,826,754 – 4,826,823 |
|---|---|
| Length | 69 |
| Sequences | 4 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 66.67 |
| Shannon entropy | 0.54807 |
| G+C content | 0.36096 |
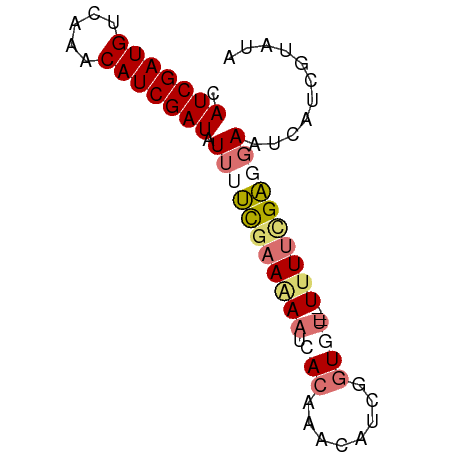
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -10.84 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
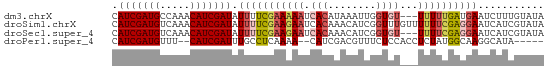
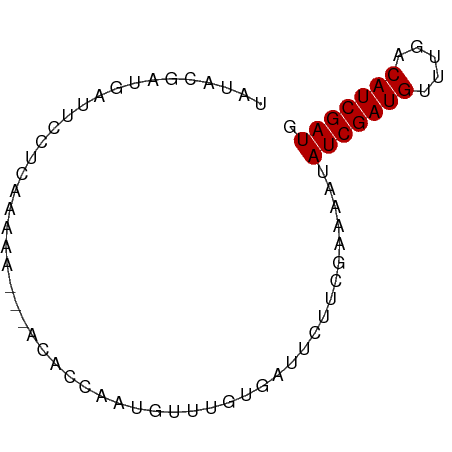
>dm3.chrX 4826754 69 + 22422827 CAUCGAUGCCAAACAUCGAUAUUUUCGAAAAAUCACAUAAAUUGGUGU---UUUUUGAUGAAUCUUUGUAUA .(((((((.....)))))))....((((((((.(((........))))---))))))).............. ( -15.00, z-score = -1.94, R) >droSim1.chrX 3732553 72 + 17042790 CAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUUUGUUUUUUCGAGGAAUCAUCGUAUA .(((((((.....))))))).(((((((((((..((((((....)))))))))))))))))........... ( -22.50, z-score = -4.10, R) >droSec1.super_4 4259802 69 - 6179234 CAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUGU---UUUUCGAGGAAUCAUCGUAUA .(((((((.....))))))).(((((((((((.(((........))))---))))))))))........... ( -20.70, z-score = -3.44, R) >droPer1.super_4 2834029 63 + 7162766 CAUCGAUGUUU--CAUCGAUUUGCCUCAAAA--CAUCGACGUUUCUCCACCUCUAUGGCAAGGCAUA----- .(((((((...--))))))).(((((.....--(((.((.((......)).)).)))...)))))..----- ( -14.30, z-score = -2.38, R) >consensus CAUCGAUGUCAAACAUCGAUAUUUUCGAAAAAUCACAAACAUCGGUGU___UUUUCGAGGAAUCAUCGUAUA .(((((((.....))))))).((((((((((..(((........)))....))))))))))........... (-10.84 = -12.28 + 1.44)
| Location | 4,826,754 – 4,826,823 |
|---|---|
| Length | 69 |
| Sequences | 4 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 66.67 |
| Shannon entropy | 0.54807 |
| G+C content | 0.36096 |
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -7.82 |
| Energy contribution | -7.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
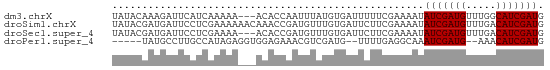
>dm3.chrX 4826754 69 - 22422827 UAUACAAAGAUUCAUCAAAAA---ACACCAAUUUAUGUGAUUUUUCGAAAAUAUCGAUGUUUGGCAUCGAUG ..............((.((((---((((........))).))))).))...((((((((.....)))))))) ( -12.30, z-score = -1.34, R) >droSim1.chrX 3732553 72 - 17042790 UAUACGAUGAUUCCUCGAAAAAACAAACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUG ....(((((.(((...)))....(((((((((((((.(((....))).))))))))..))))).)))))... ( -17.40, z-score = -2.30, R) >droSec1.super_4 4259802 69 + 6179234 UAUACGAUGAUUCCUCGAAAA---ACACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUG ....(((((.(((...)))((---(((.((((((((.(((....))).)))))))).)))))..)))))... ( -18.00, z-score = -2.38, R) >droPer1.super_4 2834029 63 - 7162766 -----UAUGCCUUGCCAUAGAGGUGGAGAAACGUCGAUG--UUUUGAGGCAAAUCGAUG--AAACAUCGAUG -----..((((((.((((....)))).((((((....))--)))))))))).(((((((--...))))))). ( -19.90, z-score = -3.20, R) >consensus UAUACGAUGAUUCCUCAAAAA___ACACCAAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUG ....................................................(((((((.....))))))). ( -7.82 = -7.82 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:28 2011