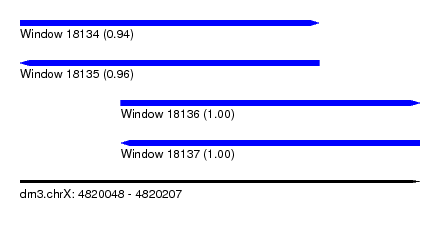
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,820,048 – 4,820,207 |
| Length | 159 |
| Max. P | 0.998452 |

| Location | 4,820,048 – 4,820,167 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.27 |
| Shannon entropy | 0.16070 |
| G+C content | 0.58888 |
| Mean single sequence MFE | -45.99 |
| Consensus MFE | -39.01 |
| Energy contribution | -39.57 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4820048 119 + 22422827 AAGGGCCCCCCAGUAUCCUGUAGUUGGACUCAAACAAGUCCCUCC-UUCCGCUUGACUGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGU ..((((....((((.......((..(((((......)))))...)-)........))))..(((((((((((..((((..((((((...))))))..))))..))))))))))))))).. ( -45.06, z-score = -3.48, R) >droSim1.chrX 3726708 118 + 17042790 AAGGGCCCC--AGCGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGU .(((((...--...)))))((((..(((((......))))).......))))...((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..)))) ( -46.50, z-score = -1.55, R) >droSec1.super_4 4252413 118 - 6179234 AAGGGCCCC--AGUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGU .(((((...--...)))))((((..(((((......))))).......))))...((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..)))) ( -46.40, z-score = -1.79, R) >consensus AAGGGCCCC__AGUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGU ...(((........(((..((((..(((((......))))).......))))..)))....((.((((..((..(((((.((((((...)))))).)))))..))..)))).)))))... (-39.01 = -39.57 + 0.56)

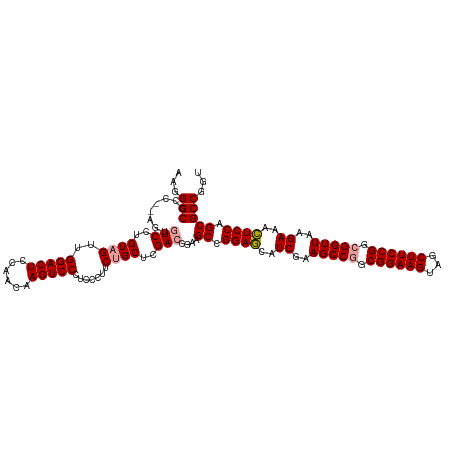
| Location | 4,820,048 – 4,820,167 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.27 |
| Shannon entropy | 0.16070 |
| G+C content | 0.58888 |
| Mean single sequence MFE | -51.87 |
| Consensus MFE | -45.65 |
| Energy contribution | -44.77 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4820048 119 - 22422827 ACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCAGUCAAGCGGAA-GGAGGGACUUGUUUGAGUCCAACUACAGGAUACUGGGGGGCCCUU ...((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))..........((-((..((((((....))))))..(..(((....)))..)...)))) ( -50.90, z-score = -3.31, R) >droSim1.chrX 3726708 118 - 17042790 ACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACGCU--GGGGCCCUU .(((((((((((((..(((..(((.((((((...)))))).)))((((..(((((.(((....)))))))))))).)))..))))(((((...)))))..)))..))))--))....... ( -53.10, z-score = -1.73, R) >droSec1.super_4 4252413 118 + 6179234 ACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACACU--GGGGCCCUU (((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).......(((((..((((((....))))))..(..(((....))--)..)))))) ( -51.60, z-score = -1.75, R) >consensus ACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACACU__GGGGCCCUU (((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).)))))...........(((((.(((...((.((((.......)))).))..))).))))) (-45.65 = -44.77 + -0.88)

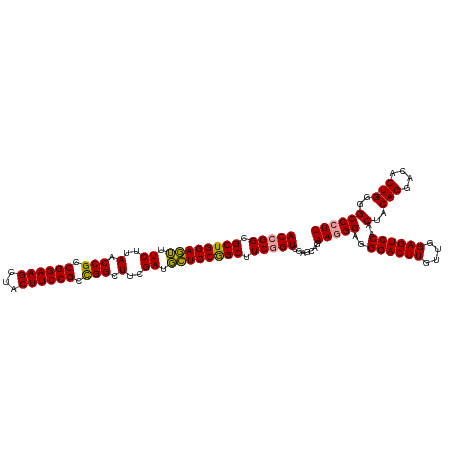
| Location | 4,820,088 – 4,820,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Shannon entropy | 0.17601 |
| G+C content | 0.58485 |
| Mean single sequence MFE | -55.87 |
| Consensus MFE | -45.30 |
| Energy contribution | -46.97 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.91 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4820088 119 + 22422827 -CCUCCUUCCGCUUGACUGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCAGGAGCGAACUUGUUGGAGUCAAACUACAGGC -.(((((..(((((.((.(.((((((((((((..((((..((((((...))))))..))))..)))))))))))).).)).)))))..)))))....(((((.(........).))))). ( -53.20, z-score = -4.60, R) >droSim1.chrX 3726746 120 + 17042790 CCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGAGUCAAACUACAGGC ....(((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))).....((((((.(........).)))))) ( -57.50, z-score = -3.54, R) >droSec1.super_4 4252451 120 - 6179234 CCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGAUUGUUGGAGUCAAACUUCAGGC .((((((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..)))))))))...)))....((((((.....)))))).. ( -56.90, z-score = -3.57, R) >consensus CCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGAGUCAAACUACAGGC ....(((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))).....((((((.(........).)))))) (-45.30 = -46.97 + 1.67)


| Location | 4,820,088 – 4,820,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Shannon entropy | 0.17601 |
| G+C content | 0.58485 |
| Mean single sequence MFE | -53.40 |
| Consensus MFE | -47.73 |
| Energy contribution | -46.30 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.998452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4820088 119 - 22422827 GCCUGUAGUUUGACUCCAACAAGUUCGCUCCUGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCAGUCAAGCGGAAGGAGG- ...........((((......))))..(((((..(((((.((.((((((((((((((..(.(((.((((((...)))))).))).)..)))))))))))))).)).)))))..))))).- ( -52.60, z-score = -4.49, R) >droSim1.chrX 3726746 120 - 17042790 GCCUGUAGUUUGACUCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGG .(((((((((((.......)))))).))((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))))))) ( -55.80, z-score = -3.73, R) >droSec1.super_4 4252451 120 + 6179234 GCCUGAAGUUUGACUCCAACAAUCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGG ((..((...(((....)))...))..))((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))))... ( -51.80, z-score = -3.06, R) >consensus GCCUGUAGUUUGACUCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGG ((..((...(((....)))...))..))((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))))... (-47.73 = -46.30 + -1.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:24 2011