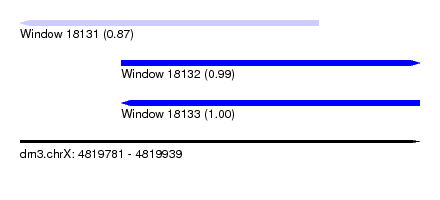
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,819,781 – 4,819,939 |
| Length | 158 |
| Max. P | 0.997506 |

| Location | 4,819,781 – 4,819,899 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.55 |
| Shannon entropy | 0.18366 |
| G+C content | 0.59957 |
| Mean single sequence MFE | -50.67 |
| Consensus MFE | -40.66 |
| Energy contribution | -39.67 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4819781 118 - 22422827 UCACAGGCGCUGGAAAAUCUUAACCGCC-GGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCAGUCGAGCGGAA-GGAGGGACUUGUUUGAGUCCAACUACAGGAUACUGGGGGGCC .....((((((((((((((..(.(((.(-(((((...)))))).))).)..)))))))))))))).(((...(....-)...((((((....))))))..(..(((....)))..)))). ( -50.70, z-score = -3.52, R) >droSim1.chrX 3726445 117 - 17042790 UCACCGGCGCUGGAGUUUCUUAACCGCC-GGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACGCUGGGGCC-- ...(((((((((((((..(((..(((.(-(((((...)))))).)))((((..(((((.(((....)))))))))))).)))..))))(((((...)))))..)))..))))))....-- ( -53.10, z-score = -1.99, R) >droSec1.super_4 4252150 117 + 6179234 UCACCGGCGCUGGAGUUUCUUAACCGCCGGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCC-ACUACAGGACACUGGGGCC-- .....(((.(..((((..(((..(((.(((((......))))).)))((((..(((((.(((....)))))))))))).)))..))).......((((-......)))).)..).)))-- ( -48.20, z-score = -0.84, R) >consensus UCACCGGCGCUGGAGUUUCUUAACCGCC_GGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACACUGGGGCC__ (((((((.((((((((.((..(.(((.(.(((((...)))))).))).)..)).)))))))).))))).))...........((((((....))))))..(..(((....)))..).... (-40.66 = -39.67 + -0.99)

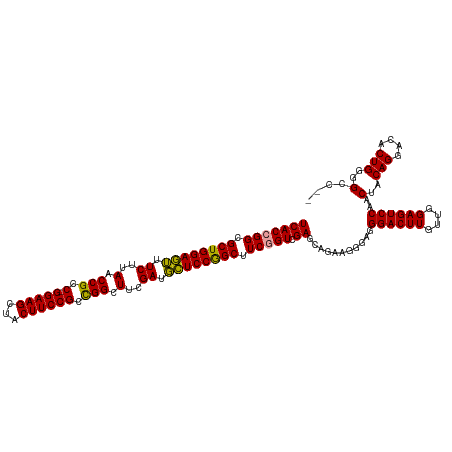
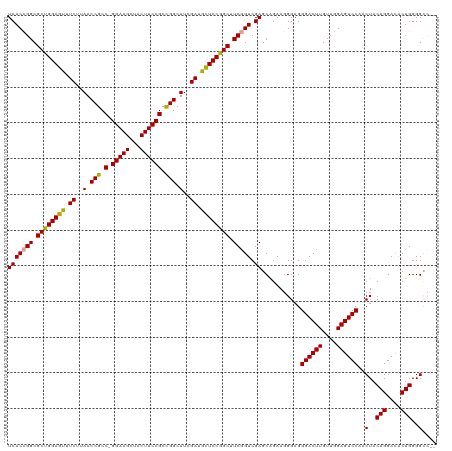
| Location | 4,819,821 – 4,819,939 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.30 |
| Shannon entropy | 0.16070 |
| G+C content | 0.57689 |
| Mean single sequence MFE | -52.80 |
| Consensus MFE | -43.60 |
| Energy contribution | -45.27 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4819821 118 + 22422827 -CCUUCCGCUCGACUGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCC-GGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCAGGAGCGAACUUGUUGGAGUCAAACUACAGGCUA -(((..((((..((.(.((((((((((((..((((..((((((...)))))-)..))))..)))))))))))).).))..))))..)))......(((((.(........).)))))... ( -46.10, z-score = -2.60, R) >droSim1.chrX 3726483 119 + 17042790 CCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCC-GGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGAGUCAAACUACAGGCUA .(((((((((..((((..((.((((..((..(((((.((((((...)))))-).)))))..))..)))).))..))))..)))))))))....(((((((.(........).))))))). ( -60.00, z-score = -4.45, R) >droSec1.super_4 4252187 120 - 6179234 CCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGAUUGUUGGAGUCAAACUUCAGGCUA .(((((((((..((((..((.((((..((..(((((.((((((...))).))).)))))..))..)))).))..))))..)))))))))(((.......((((((.....))))))))). ( -52.31, z-score = -2.43, R) >consensus CCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCC_GGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGAGUCAAACUACAGGCUA .(((((((((..((((..((.((((..((..(((((.((((((...))))).).)))))..))..)))).))..))))..)))))))))....(((((((.(........).))))))). (-43.60 = -45.27 + 1.67)


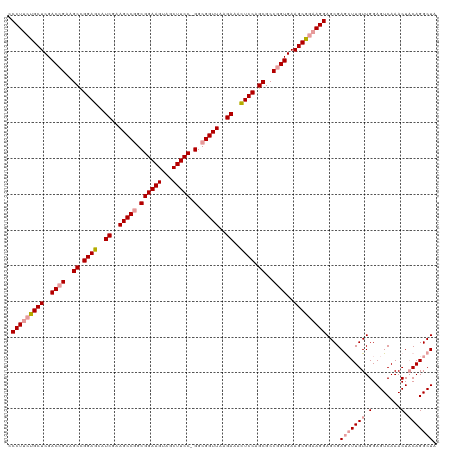
| Location | 4,819,821 – 4,819,939 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.30 |
| Shannon entropy | 0.16070 |
| G+C content | 0.57689 |
| Mean single sequence MFE | -51.70 |
| Consensus MFE | -43.09 |
| Energy contribution | -42.10 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4819821 118 - 22422827 UAGCCUGUAGUUUGACUCCAACAAGUUCGCUCCUGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCC-GGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCAGUCGAGCGGAAGG- ...(((...((((((((..........(((.....))).......((((((((((((((..(.(((.(-(((((...)))))).))).)..))))))))))))))))))))))...)))- ( -50.80, z-score = -4.30, R) >droSim1.chrX 3726483 119 - 17042790 UAGCCUGUAGUUUGACUCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCC-GGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGG .(((....((((((.......)))))).)))(((((.((((.(((((.((((((((.((..(.(((.(-(((((...)))))).))).)..)).)))))))).))))).)))).))))). ( -54.10, z-score = -3.98, R) >droSec1.super_4 4252187 120 + 6179234 UAGCCUGAAGUUUGACUCCAACAAUCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGG .(((..((...(((....)))...))..)))(((((.((((.(((((.((((((((.((..(.(((.(((((......))))).))).)..)).)))))))).))))).)))).))))). ( -50.20, z-score = -2.87, R) >consensus UAGCCUGUAGUUUGACUCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCC_GGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGG .(((..((...(((....)))...))..)))(((((.((((.(((((.((((((((.((..(.(((.(.(((((...)))))).))).)..)).)))))))).))))).)))).))))). (-43.09 = -42.10 + -0.99)

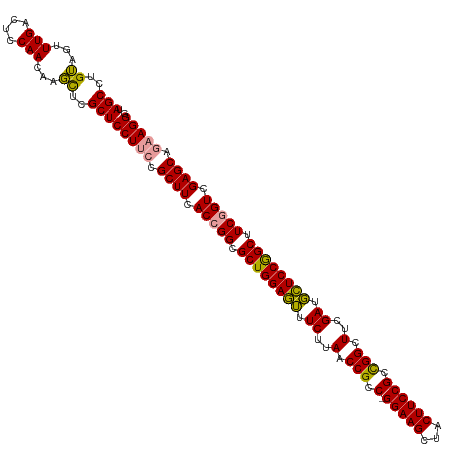
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:21 2011