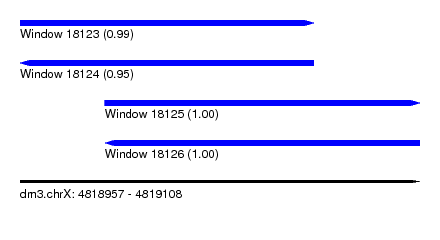
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,818,957 – 4,819,108 |
| Length | 151 |
| Max. P | 0.999570 |

| Location | 4,818,957 – 4,819,068 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.17 |
| Shannon entropy | 0.41040 |
| G+C content | 0.58621 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -28.32 |
| Energy contribution | -29.67 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818957 111 + 22422827 --------GGGCCCCCCAGUAUCCUGUAGUUGGACUCAAACAAGUCCCUCC-UUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGC --------((((....(((....))).....(((((......)))))....-....)))).......(((((((((((..((((..((((((...))))))..))))..))))))))))) ( -39.50, z-score = -2.72, R) >droSim1.chrX 13316169 120 - 17042790 GUUGAAUGAGGCGCCCUGGUAGCCUGUAGUUUGACUCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGC ((((..(((((((....((.(((....((((((.......)))))).)))))...))))))).))))((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))) ( -47.40, z-score = -2.50, R) >droSec1.super_4 4252947 110 - 6179234 ----------GGGCCCCAGUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGC ----------.(((....(.(((..((((..(((((......))))).......))))..))))...)))((((..((..(((((.((((((...)))))).)))))..))..))))... ( -42.10, z-score = -1.83, R) >consensus _________GGCGCCCCAGUAUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCCGCUCGACCGACGCUGGAGAAUCAAAGCCGGCGGAAGUAACUUCCGGCGGUUAAGAUACUCCAGC ........((((....(((....))).....(((((......))))).........)))).......((((((((.((..(((((.((((((...)))))).)))))..)).)))))))) (-28.32 = -29.67 + 1.35)


| Location | 4,818,957 – 4,819,068 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.17 |
| Shannon entropy | 0.41040 |
| G+C content | 0.58621 |
| Mean single sequence MFE | -46.27 |
| Consensus MFE | -32.11 |
| Energy contribution | -32.90 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818957 111 - 22422827 GCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAA-GGAGGGACUUGUUUGAGUCCAACUACAGGAUACUGGGGGGCCC-------- (((((((((((..(.(((.((((((...)))))).))).)..))))))))))).((((.....)))).-((..((((((....))))))..(..(((....)))..)..)).-------- ( -45.30, z-score = -2.50, R) >droSim1.chrX 13316169 120 + 17042790 GCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGAGUCAAACUACAGGCUACCAGGGCGCCUCAUUCAAC ...((((..((..(((((.((((((...)))))).)))))..))..)))).((((((((...((.......)).(((((((.(........).))))))))))..))))).......... ( -48.00, z-score = -1.69, R) >droSec1.super_4 4252947 110 + 6179234 GCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACACUGGGGCCC---------- (((.((((..(((..(((.((((((...)))))).)))((((..(((((.(((....)))))))))))).)))..)))).(..(.((((.......)))).)..))))..---------- ( -45.50, z-score = -1.24, R) >consensus GCUGGAGAAUCUUAACCGCCGGAAGUAACUUCCGCCGGCUUAGAUACUCCAGCGUCGGUCGAGCGGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGAUACUGGGGCGCC_________ ((((((((.((....(((.((((((...)))))).)))....)).))))))))....................((((((....))))))..((.(((....))).))............. (-32.11 = -32.90 + 0.79)

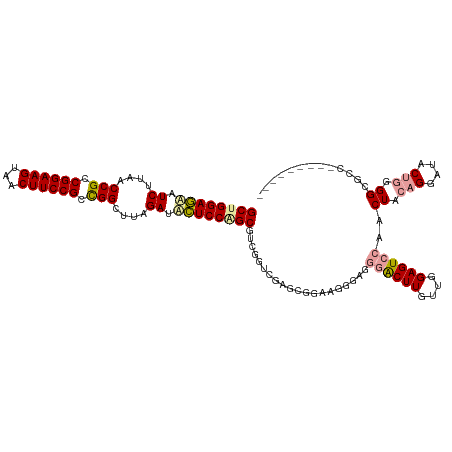
| Location | 4,818,989 – 4,819,108 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.38954 |
| G+C content | 0.58487 |
| Mean single sequence MFE | -56.67 |
| Consensus MFE | -50.14 |
| Energy contribution | -50.27 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818989 119 + 22422827 CAAGUCCCUCC-UUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCGGGAGCGAACUUGUUGGAGUCAAACU (((((.(((((-(..((((..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))..))))..))))).).)))))............. ( -56.80, z-score = -5.32, R) >droSim1.chrX 13316209 120 - 17042790 CAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACU .((((((..((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).)))))).))).)))(((((...))))). ( -58.80, z-score = -3.77, R) >droSec1.super_4 4252977 120 - 6179234 CAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGAUUGUUGGAGUCAAACU ..(((((((((.(((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))))).).)))).............. ( -54.40, z-score = -3.37, R) >consensus CAAGUCCCUCCCUUCCGCUCGACCGACGCUGGAGAAUCAAAGCCGGCGGAAGUAACUUCCGGCGGUUAAGAUACUCCAGCGCCGGUGAAGCGGAAGGAGCGAACUUGUUGGAGUCAAACU ((((((.((((.(((((((..((((..((((((((.((..(((((.((((((...)))))).)))))..)).))))))))..))))..))))))))))).)).))))............. (-50.14 = -50.27 + 0.13)


| Location | 4,818,989 – 4,819,108 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.38954 |
| G+C content | 0.58487 |
| Mean single sequence MFE | -59.47 |
| Consensus MFE | -48.57 |
| Energy contribution | -50.80 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -5.06 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818989 119 - 22422827 AGUUUGACUCCAACAAGUUCGCUCCCGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAA-GGAGGGACUUG .............(((((((.((((...(((((.(((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).)))))...-))))))))))) ( -63.30, z-score = -6.86, R) >droSim1.chrX 13316209 120 + 17042790 AGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUG .(((((...)))))(((..(((((.(((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))))).)..))). ( -60.20, z-score = -4.39, R) >droSec1.super_4 4252977 120 + 6179234 AGUUUGACUCCAACAAUCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUG .............(((((((..((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).)))))).)))).))) ( -54.90, z-score = -3.92, R) >consensus AGUUUGACUCCAACAAGUUCGCUCCUUCCGCUUCACCGGCGCUGGAGAAUCUUAACCGCCGGAAGUAACUUCCGCCGGCUUAGAUACUCCAGCGUCGGUCGAGCGGAAGGGAGGGACUUG .............(((((((.((((((((((((.(((((.((((((((.((....(((.((((((...)))))).)))....)).)))))))).))))).)))))))))).))))))))) (-48.57 = -50.80 + 2.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:16 2011