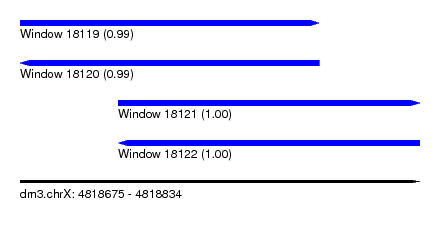
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,818,675 – 4,818,834 |
| Length | 159 |
| Max. P | 0.999518 |

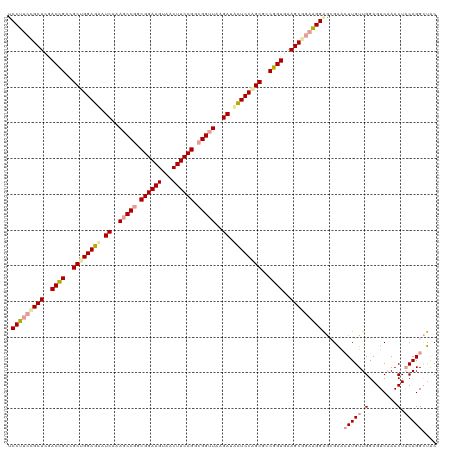
| Location | 4,818,675 – 4,818,794 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.37424 |
| G+C content | 0.59211 |
| Mean single sequence MFE | -46.03 |
| Consensus MFE | -35.24 |
| Energy contribution | -35.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818675 119 + 22422827 GGCCCCCCAGUAUCCUGUAGUUGGACUCAAACAAGUCCCUCC-UUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAA (((....(((....))).....(((((......)))))....-....)))..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))... ( -44.60, z-score = -3.66, R) >droSim1.chrX 13315912 120 - 17042790 GGCGCCCUGGUAGCCUGUAGUUUGACUCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGA ((((....((.(((....((((((.......)))))).)))))...))))(((((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)) ( -47.30, z-score = -2.01, R) >droSec1.super_4 4252681 119 - 6179234 -GGGCCCCAGUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAA -((((......)))).((((..(((((......))))).......))))((.((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..)))))). ( -46.20, z-score = -1.44, R) >consensus GGCGCCCCAGUAUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCCGCUCGACCGACGCUGGAGAAUCAAAGCCGGCGGAAGUAACUUCCGGCGGUUAAGAUACUCCAGCGCCGGUGAA ......................(((((......)))))..............((((..((((((((.((..(((((.((((((...)))))).)))))..)).))))))))..))))... (-35.24 = -35.80 + 0.56)


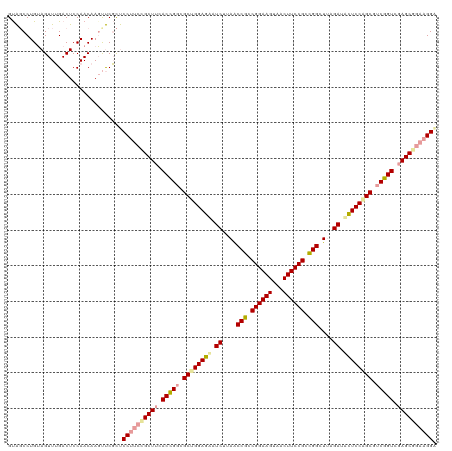
| Location | 4,818,675 – 4,818,794 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.37424 |
| G+C content | 0.59211 |
| Mean single sequence MFE | -51.23 |
| Consensus MFE | -38.12 |
| Energy contribution | -38.80 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818675 119 - 22422827 UUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAA-GGAGGGACUUGUUUGAGUCCAACUACAGGAUACUGGGGGGCC .(((((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).))((....-....((((((....))))))..(..(((....)))..).)). ( -55.10, z-score = -4.54, R) >droSim1.chrX 13315912 120 + 17042790 UCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGAGUCAAACUACAGGCUACCAGGGCGCC ............((((..((..(((((.((((((...)))))).)))))..))..)))).((((((((...((.......)).(((((((.(........).))))))))))..))))). ( -48.00, z-score = -1.19, R) >droSec1.super_4 4252681 119 + 6179234 UUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACACUGGGGCCC- ((((((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).))).....(((..((((((....))))))..(..(((....)))..))))- ( -50.60, z-score = -1.50, R) >consensus UUCACCGGCGCUGGAGAAUCUUAACCGCCGGAAGUAACUUCCGCCGGCUUAGAUACUCCAGCGUCGGUCGAGCGGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGAUACUGGGGCGCC ...(((((.((((((((.((....(((.((((((...)))))).)))....)).)))))))).)))))..............((((((....))))))..((.(((....))).)).... (-38.12 = -38.80 + 0.68)

| Location | 4,818,714 – 4,818,834 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.38189 |
| G+C content | 0.58333 |
| Mean single sequence MFE | -55.30 |
| Consensus MFE | -48.31 |
| Energy contribution | -49.77 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818714 120 + 22422827 UCCUUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCGGGAGCGAACUUGUUGGAGUCAAACUACAGGCUAC ((((..((((..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))..))))..)))).....(((((.(........).))))).... ( -52.30, z-score = -4.09, R) >droSim1.chrX 13315952 120 - 17042790 UCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACGC ((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))))..((((((....))))))............. ( -57.50, z-score = -3.50, R) >droSec1.super_4 4252720 120 - 6179234 CCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGAUUGUUGGAGUCAAACUUCAGGCUAC .(((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..)))))))))(((.......((((((.....))))))))).. ( -56.11, z-score = -3.75, R) >consensus UCCUUCCGCUCGACCGACGCUGGAGAAUCAAAGCCGGCGGAAGUAACUUCCGGCGGUUAAGAUACUCCAGCGCCGGUGAAGCGGAAGGAGCGAACUUGUUGGAGUCAAACUACAGGCUAC .(((((((((..((((..((((((((.((..(((((.((((((...)))))).)))))..)).))))))))..))))..)))))))))......(((((.(........).))))).... (-48.31 = -49.77 + 1.45)


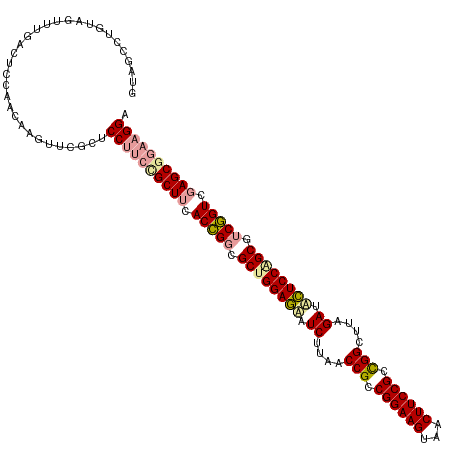
| Location | 4,818,714 – 4,818,834 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.38189 |
| G+C content | 0.58333 |
| Mean single sequence MFE | -54.73 |
| Consensus MFE | -45.51 |
| Energy contribution | -46.63 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -4.16 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4818714 120 - 22422827 GUAGCCUGUAGUUUGACUCCAACAAGUUCGCUCCCGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAAGGA ...(((.(.(((..((((......)))).))).).)))((((.(((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).))))....... ( -55.40, z-score = -5.02, R) >droSim1.chrX 13315952 120 + 17042790 GCGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGA .............(((((......)))))...(((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))). ( -56.80, z-score = -3.75, R) >droSec1.super_4 4252720 120 + 6179234 GUAGCCUGAAGUUUGACUCCAACAAUCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGG ..(((..((...(((....)))...))..)))(((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))). ( -52.00, z-score = -3.70, R) >consensus GUAGCCUGUAGUUUGACUCCAACAAGUUCGCUCCUUCCGCUUCACCGGCGCUGGAGAAUCUUAACCGCCGGAAGUAACUUCCGCCGGCUUAGAUACUCCAGCGUCGGUCGAGCGGAAGGA ................................((((((((((.(((((.((((((((.((....(((.((((((...)))))).)))....)).)))))))).))))).)))))))))). (-45.51 = -46.63 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:13 2011