
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,817,857 – 4,818,021 |
| Length | 164 |
| Max. P | 0.999856 |

| Location | 4,817,857 – 4,818,021 |
|---|---|
| Length | 164 |
| Sequences | 3 |
| Columns | 165 |
| Reading direction | forward |
| Mean pairwise identity | 64.85 |
| Shannon entropy | 0.49928 |
| G+C content | 0.55169 |
| Mean single sequence MFE | -65.47 |
| Consensus MFE | -54.09 |
| Energy contribution | -54.33 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
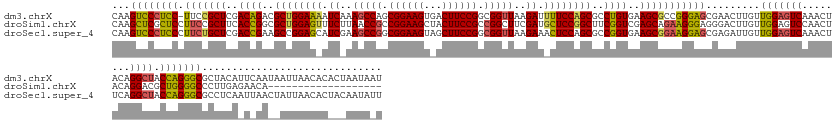
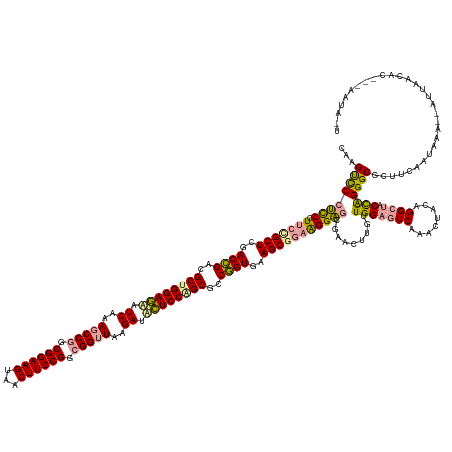
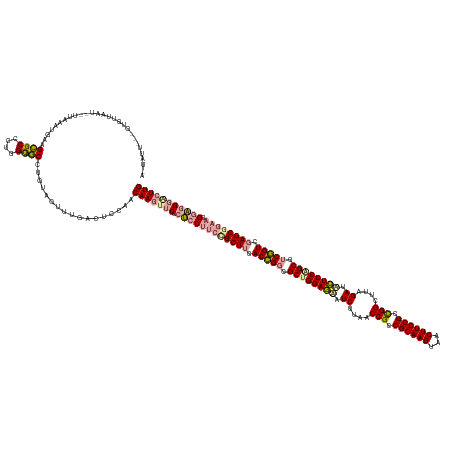
>dm3.chrX 4817857 164 + 22422827 CAAGUCCCUCC-UUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCGGGAGCGAACUUGUUGGAGUCAAACUACAGGCUACCAGGGCGCUACAUUCAAUAAUUAACACACUAAUAAU (((((.(((((-(..((((..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))..))))..))))).).)))))((((((((........)))).))))................................. ( -63.90, z-score = -4.97, R) >droSim1.chrX 13316209 146 - 17042790 CAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACGCUGGGGCCCUUGAGAACA------------------- ....(((...(((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).)))))(((((.((..(..(.((((.......)))).)..)))))))))))....------------------- ( -67.90, z-score = -2.85, R) >droSec1.super_4 4252445 165 - 6179234 CAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGAUUGUUGGAGUCAAACUUCAGGCUACCAGGGCGCCUCAAUUAACUAUUAACACUACAAUAUU ...((((((((((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..)))))))))...))).....(((((((........)))).))))))).............................. ( -64.60, z-score = -3.25, R) >consensus CAAGUCCCUCCCUUCCGCUCGACCGACGCUGGAGAAUCAAAGCCGGCGGAAGUAACUUCCGGCGGUUAAGAUACUCCAGCGCCGGUGAAGCGGAAGGAGCGAACUUGUUGGAGUCAAACUACAGGCUACCAGGGCGCUUCAAUAAA__AUUAACAC___AAUA_U ...((((((((.(((((((..((((..((((((((.((..(((((.((((((...)))))).)))))..)).))))))))..))))..))))))))))).........(((((((........)))).))))))).............................. (-54.09 = -54.33 + 0.24)
| Location | 4,817,857 – 4,818,021 |
|---|---|
| Length | 164 |
| Sequences | 3 |
| Columns | 165 |
| Reading direction | reverse |
| Mean pairwise identity | 64.85 |
| Shannon entropy | 0.49928 |
| G+C content | 0.55169 |
| Mean single sequence MFE | -69.30 |
| Consensus MFE | -50.23 |
| Energy contribution | -51.80 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
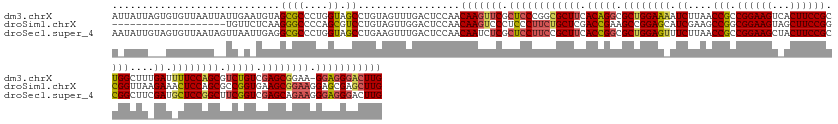
>dm3.chrX 4817857 164 - 22422827 AUUAUUAGUGUGUUAAUUAUUGAAUGUAGCGCCCUGGUAGCCUGUAGUUUGACUCCAACAAGUUCGCUCCCGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAA-GGAGGGACUUG .(((((((.(((((((((....))).)))))).)))))))..................(((((((.((((...(((((.(((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).)))))...-))))))))))) ( -72.40, z-score = -5.87, R) >droSim1.chrX 13316209 146 + 17042790 -------------------UGUUCUCAAGGGCCCCAGCGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUG -------------------.........(((..(((((........)))))..)))..((((..(((((.(((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))))).)..)))) ( -67.90, z-score = -3.54, R) >droSec1.super_4 4252445 165 + 6179234 AAUAUUGUAGUGUUAAUAGUUAAUUGAGGCGCCCUGGUAGCCUGAAGUUUGACUCCAACAAUCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUG ..........((((...((((((((.((((((....)).)))).))..))))))..)))).((((..((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).)))))).)))).... ( -67.60, z-score = -3.36, R) >consensus A_UAUU___GUGUUAAU__UUAAAUGAAGCGCCCUGGUAGCCUGUAGUUUGACUCCAACAAGUUCGCUCCUUCCGCUUCACCGGCGCUGGAGAAUCUUAACCGCCGGAAGUAACUUCCGCCGGCUUAGAUACUCCAGCGUCGGUCGAGCGGAAGGGAGGGACUUG ............................((((....)).)).................(((((((.((((((((((((.(((((.((((((((.((....(((.((((((...)))))).)))....)).)))))))).))))).)))))))))).))))))))) (-50.23 = -51.80 + 1.57)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:07 2011