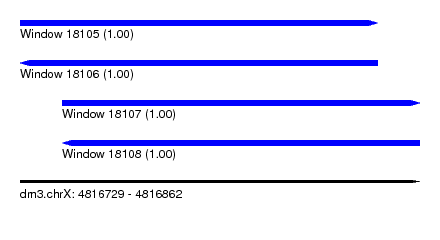
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,816,729 – 4,816,862 |
| Length | 133 |
| Max. P | 0.999113 |

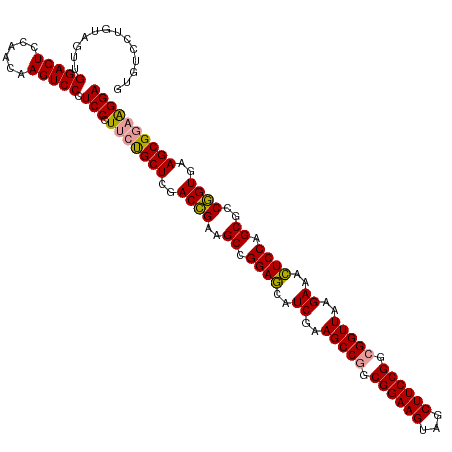
| Location | 4,816,729 – 4,816,848 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Shannon entropy | 0.16070 |
| G+C content | 0.58485 |
| Mean single sequence MFE | -55.90 |
| Consensus MFE | -49.96 |
| Energy contribution | -49.97 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.34 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.998995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4816729 119 + 22422827 GUAUCCUGUAGUUGGACUCAAACAAGUCCCUCC-UUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCGGGA ...(((((.....(((((......)))))....-...((((..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))..)))).))))) ( -54.10, z-score = -5.22, R) >droSim1.chrX 3726452 120 + 17042790 GCGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGA .............(((((......)))))...(((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))). ( -56.80, z-score = -3.75, R) >droSec1.super_4 4250841 120 - 6179234 GUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGA .............(((((......)))))...(((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))). ( -56.80, z-score = -4.05, R) >consensus GUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGA .............(((((......)))))...(((((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))). (-49.96 = -49.97 + 0.01)

| Location | 4,816,729 – 4,816,848 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Shannon entropy | 0.16070 |
| G+C content | 0.58485 |
| Mean single sequence MFE | -58.50 |
| Consensus MFE | -53.47 |
| Energy contribution | -53.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.63 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4816729 119 - 22422827 UCCCGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAA-GGAGGGACUUGUUUGAGUCCAACUACAGGAUAC (((.(.(((((.(((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).)))))...-((..((((((....))))))..)).).)))... ( -60.50, z-score = -6.39, R) >droSim1.chrX 3726452 120 - 17042790 UCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACGC ((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))))..((((((....))))))............. ( -57.50, z-score = -3.50, R) >droSec1.super_4 4250841 120 + 6179234 UCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACAC ((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))))..((((((....))))))............. ( -57.50, z-score = -3.99, R) >consensus UCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACAC ((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))))))..((((((....))))))............. (-53.47 = -53.03 + -0.44)

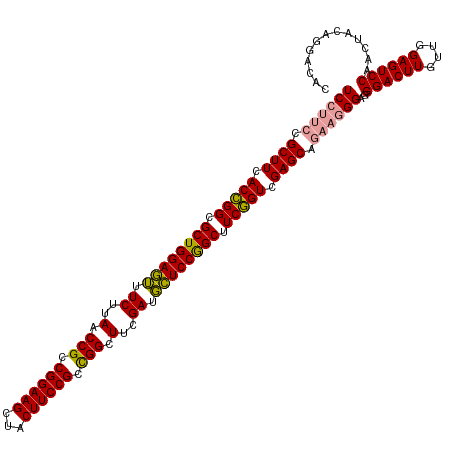
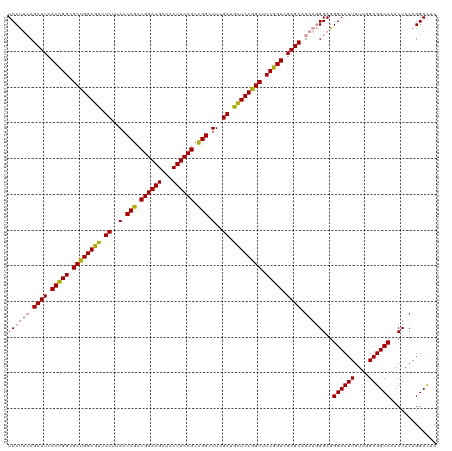
| Location | 4,816,743 – 4,816,862 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Shannon entropy | 0.16070 |
| G+C content | 0.58765 |
| Mean single sequence MFE | -63.80 |
| Consensus MFE | -58.41 |
| Energy contribution | -58.53 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -6.08 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4816743 119 + 22422827 GACUCAAACAAGUCCCUCC-UUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCGGGAGCGAACUUGUUGGA ...((.(((((((.(((((-(..((((..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))..))))..))))).).))))))).)) ( -60.20, z-score = -6.34, R) >droSim1.chrX 3726466 120 + 17042790 GACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGA ...(((((((((..(((((.(((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))))).)..))))))))) ( -67.60, z-score = -6.26, R) >droSec1.super_4 4250855 120 - 6179234 GACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGAUUGUUGGA ...((((((((.((.((((.(((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))))).))..)))))))) ( -63.60, z-score = -5.64, R) >consensus GACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGA ...((((((((((.(((((.(((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..))))))))))).).)))))))))) (-58.41 = -58.53 + 0.12)


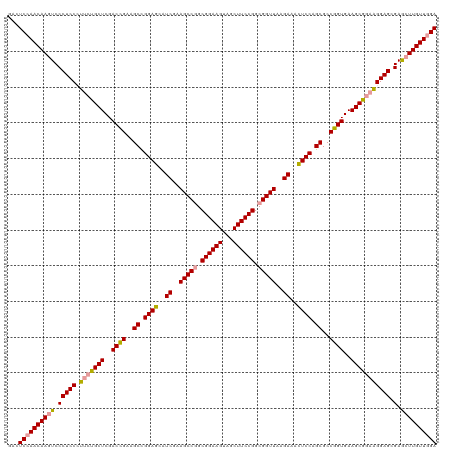
| Location | 4,816,743 – 4,816,862 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Shannon entropy | 0.16070 |
| G+C content | 0.58765 |
| Mean single sequence MFE | -66.90 |
| Consensus MFE | -61.34 |
| Energy contribution | -60.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -6.73 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4816743 119 - 22422827 UCCAACAAGUUCGCUCCCGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAA-GGAGGGACUUGUUUGAGUC ((.(((((((((.((((...(((((.(((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).)))))...-))))))))))))).))... ( -67.50, z-score = -7.83, R) >droSim1.chrX 3726466 120 - 17042790 UCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUC ((((((((((((..((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).)))))).))).)))))))))... ( -67.60, z-score = -6.18, R) >droSec1.super_4 4250855 120 + 6179234 UCCAACAAUCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUC ((((((((((((..((((((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).)))))).)))).))))))))... ( -65.60, z-score = -6.19, R) >consensus UCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUC (((((((((.((.((((((.(((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).).)))))).)))))))))))... (-61.34 = -60.90 + -0.44)


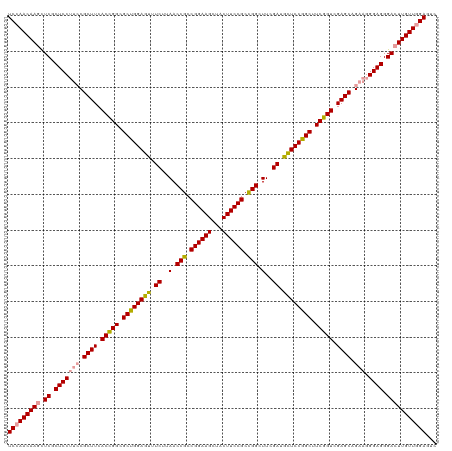
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:01 2011