
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,805,564 – 4,805,679 |
| Length | 115 |
| Max. P | 0.767899 |

| Location | 4,805,564 – 4,805,679 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 52.83 |
| Shannon entropy | 0.90983 |
| G+C content | 0.45923 |
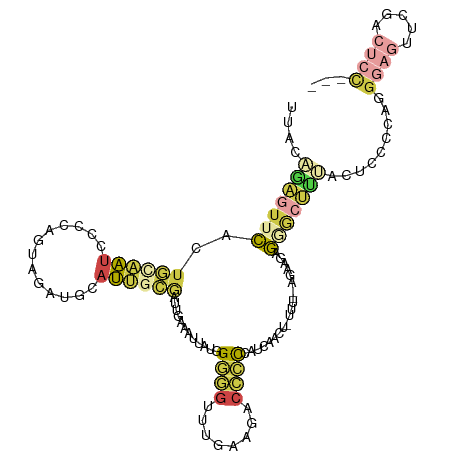
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -8.15 |
| Energy contribution | -10.27 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
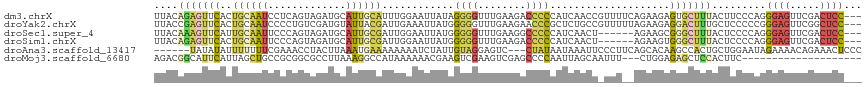
>dm3.chrX 4805564 115 + 22422827 UUACAGAGUUCACUGCAAUCCUCAGUAGAUGCAUUGCAUUUGGAAUUAUAGGGGUUUGAAGACCCCCAUCAACCGUUUUCAGAAGAGUGCUUUACUUCCCAGGGAGUUCGACUCC--- .....(((((.(((.(.......((((((.(((((.(.(((((((.....((((((....))))))..........))))))).))))))))))))......).)))..))))).--- ( -29.48, z-score = -0.83, R) >droYak2.chrX 18130776 115 - 21770863 UUACCGAGUUCACUGCAAUCCCCUGUCGAUGUAUUACGAUUGAAAUUAUGGGGGUUUGAAGAACCCGCUCUGCCGUUUUUAGAAGAGGACUUUGCUCCCCCGGGAGUUCGGCUCC--- ....((.((((....(((.(((((.(((((........)))))......))))).)))..)))).))....(((((((((....))))))...(((((....)))))..)))...--- ( -31.70, z-score = -0.16, R) >droSec1.super_4 4240550 109 - 6179234 UUACAAAGUUCAUUGCAAUUCCCAGUAGAUGCAUUGCGAUUGGAAUUAUGGGGGUUUGAAGGCCCCCAUCAACU------AGAAGCGGGCUUUACUCCCCAGGGAGUUCGACUCC--- ............(((.(((((((((((((.((.((((..((((....(((((((((....)))))))))...))------))..)))))))))))).....))))))))))....--- ( -35.50, z-score = -1.62, R) >droSim1.chrX 3715354 109 + 17042790 UUACAGAGUUCACUGCAAUUCCCAGUAGAUGCAUUGCGAUUGGAAUUAUGGGGGUUUGAAGACCCCCAUCAACU------AGAAGUGGGCUUUACUCCCCAGGGAGUUCGACUCC--- ....((((((((((.(((((((..((((.....))))....))))))(((((((((....))))))))).....------.).))))))))))(((((....)))))........--- ( -38.90, z-score = -2.77, R) >droAna3.scaffold_13417 161738 109 + 6960332 ------UAUAUAUUUUUUUCGAAACCUACUUAAAUGAAAAAAAAUCUAUUGUAGGAGUC---CUAUAAUAAAUUCCCUUCAGCACAAGCCACUGCUGGAAUAGAAAACAGAAACUCCC ------......(((((((((.............)))))))))...(((((((((...)---))))))))..(((..(((((((........)))))))...)))............. ( -17.92, z-score = -1.95, R) >droMoj3.scaffold_6680 13608044 95 - 24764193 AGACGGCAUUCAUUAGCUGCCGCGGCGCCUUAAAGGCCAUAAAAAACGAAGUCGAAGUCGAGCCCCAAUUAGCAAUUU---CUGGAGAGCUCCACUUC-------------------- .(((((((.........))))...(.(((.....))))............)))(((((.((((((((...(....)..---.))).).)))).)))))-------------------- ( -25.90, z-score = -0.91, R) >consensus UUACAGAGUUCACUGCAAUCCCCAGUAGAUGCAUUGCGAUUGAAAUUAUGGGGGUUUGAAGACCCCCAUCAACU_UUU__AGAAGAGGGCUUUACUCCCCAGGGAGUUCGACUCC___ ....(((((((..((((((.............))))))............((((((....))))))....................)))))))(((((....)))))........... ( -8.15 = -10.27 + 2.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:55 2011