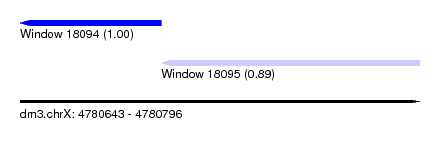
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,780,643 – 4,780,796 |
| Length | 153 |
| Max. P | 0.995915 |

| Location | 4,780,643 – 4,780,697 |
|---|---|
| Length | 54 |
| Sequences | 6 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.42908 |
| G+C content | 0.46490 |
| Mean single sequence MFE | -13.58 |
| Consensus MFE | -10.56 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
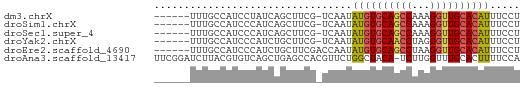
>dm3.chrX 4780643 54 - 22422827 ------UUUGCCAUCCUAUCAGCUUCG-UCAAUAUGUGCAGCCAAAGGUUGCACAUUUCCU ------.....................-.....((((((((((...))))))))))..... ( -13.50, z-score = -2.79, R) >droSim1.chrX 3692830 54 - 17042790 ------UUUGCCAUCCCAUCAGCUUCG-UCAAUAUGUGCAGCCAAAGGUUGCACAUUUCCU ------.....................-.....((((((((((...))))))))))..... ( -13.50, z-score = -2.83, R) >droSec1.super_4 4216229 54 + 6179234 ------UUUGCCAUCCCAUCAGCUUCG-UCAAUAUGUGCAGCCAAAGGUUGCACAUUUCCU ------.....................-.....((((((((((...))))))))))..... ( -13.50, z-score = -2.83, R) >droYak2.chrX 18104987 54 + 21770863 ------UUUGCCAUCCCAUCUGCUUCG-UCAAUAUGUGCAACCUAGGGUUGCACAUUUCCU ------.....................-.....((((((((((...))))))))))..... ( -13.50, z-score = -2.48, R) >droEre2.scaffold_4690 2151628 55 - 18748788 ------UUUGCCAUCCCAUCUGCUUCGACCAAUAUGUGCAGCCUAAGGUUGCACAUUUCCU ------...........................((((((((((...))))))))))..... ( -13.50, z-score = -2.47, R) >droAna3.scaffold_13417 123649 60 - 6960332 UUCGGAUCUUACGUGUCAGCUGAGCCACGUUCUGGCUACA-UCUUGGUUUGCACUUUUCCA ...(((......((((.((((.(((((.....)))))...-....)))).))))...))). ( -14.00, z-score = -1.05, R) >consensus ______UUUGCCAUCCCAUCAGCUUCG_UCAAUAUGUGCAGCCAAAGGUUGCACAUUUCCU .................................((((((((((...))))))))))..... (-10.56 = -10.70 + 0.14)
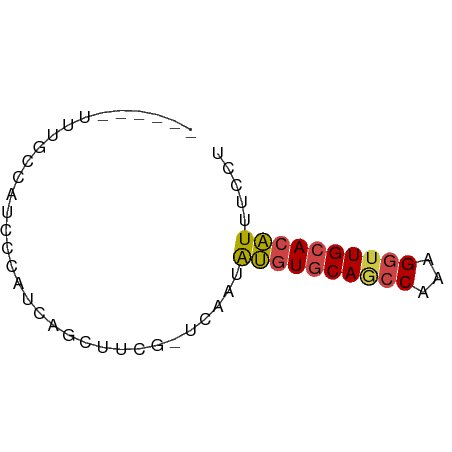
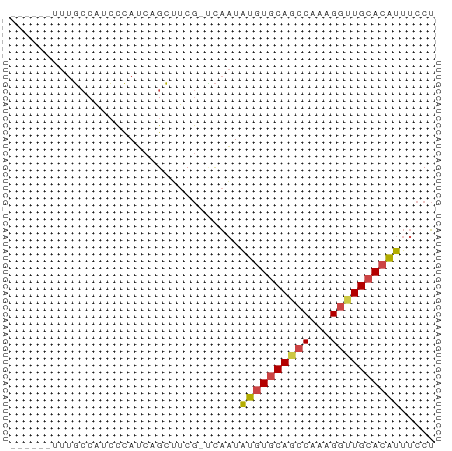
| Location | 4,780,697 – 4,780,796 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 57.97 |
| Shannon entropy | 0.71486 |
| G+C content | 0.41754 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -7.08 |
| Energy contribution | -7.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.888206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4780697 99 - 22422827 CUUCUGUACACCUUAACAUAAAUC--------CACUUUAAAUUUUU-----AUUUAAAGAUCCCAAC------AUCUCCAAGUUGCCGAUGCUACCACGCAUCCGUAUGUACGGAAUA .(((((((((..............--------..((((((((....-----))))))))........------..........(((.(((((......))))).)))))))))))).. ( -23.80, z-score = -5.85, R) >droSim1.chrX 3692884 112 - 17042790 CUUCUCUAUACCUUAGCAUAGAUUCAACUUGGCAUUUGGAGUUUUUU--GAAUUCAAAGAUUGGAGU----AGAUAUCCUAGUCGCCGUUGCUACCACGCAUCCGUGUGUGCGGAAUA ..........((.(((((...........((((.....((((((...--))))))...(((((((..----.....))).)))))))).))))).((((((....)))))).)).... ( -24.30, z-score = 0.10, R) >droSec1.super_4 4216283 112 + 6179234 CUUCUCUAUACCUUAGCAUAGCUUCAGUUUGGCAUUUUGAGUUUUUU--GAAUUUAAAGAUUGGAGU----GGAUGUCCAAGUCGCCGUUGCUACCACGCAUCCGCAUGUGCGGAAUA .............(((((.(((....))).(((.((((((((((...--))))))))))...((..(----((....)))..)))))..))))).......(((((....)))))... ( -26.00, z-score = -0.06, R) >droYak2.chrX 18105041 103 + 21770863 --------CCAUUUGGCAUUUAGAC-------UUUUUUGAAUUUUUUAAAGAUUUAAAGAUCCUAUCUACUCAAAGUCGCCGACGACGACGAUGCCACACAGCCCCAUGUGCGGCAUA --------...(((((....(((..-------.(((((((((((.....)))))))))))..))).....)))))((((....))))....(((((.((((......)))).))))). ( -25.50, z-score = -3.26, R) >droEre2.scaffold_4690 2151683 87 - 18748788 ----------------CUUUCAAG--------CACCUUAGACUUUUC-AGAAUUUCUAGAACCUAU------AUACUGCACGUCGCCGACGAUACCUCGCAUCCCUACGAGCGAAAUA ----------------.......(--------((...((.....(((-((.....)).)))....)------)...))).((((...)))).....((((.((.....)))))).... ( -10.50, z-score = 0.01, R) >consensus CUUCU_UACACCUUAGCAUAGAUUC_______CAUUUUGAAUUUUUU__GAAUUUAAAGAUCCGAGC_____AAUCUCCAAGUCGCCGAUGCUACCACGCAUCCGUAUGUGCGGAAUA ..................................((((((((((.....))))))))))..........................................(((((....)))))... ( -7.08 = -7.08 + 0.00)

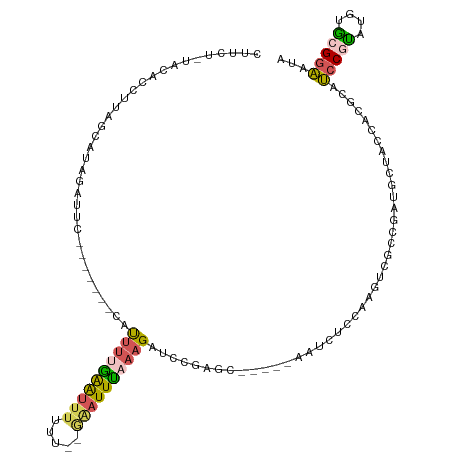
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:51 2011