
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,755,196 – 4,755,288 |
| Length | 92 |
| Max. P | 0.894217 |

| Location | 4,755,196 – 4,755,288 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.57 |
| Shannon entropy | 0.48575 |
| G+C content | 0.39727 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -14.40 |
| Energy contribution | -13.48 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4755196 92 + 22422827 AGUUUAGUUUGUCUUUC----UAAGGAUAGGA--UGGAU---AGUCCUCAAGCUCAUCUCGUUCAAGAAAACGAUGAGCUGUGGGUAUUCGGUACAAGUUU------------ .......((((((((..----..)))))))).--(((((---(.(((...((((((..(((((......)))))))))))..)))))))))..........------------ ( -23.20, z-score = -1.29, R) >droEre2.scaffold_4690 2124087 110 + 18748788 CAUUUUGUUUGACUUCCUUACGUUAAGCGGUAAGUAGG---CAGCACUCAAGCUUAUCUCGUUCAAGAAAAUGAUGAGCUGUGGAAAUUCGGGACAAGUAGUCCCUCACAUUU .....((((.(.(((.(((((........))))).)))---))))).((.((((((((...((....))...))))))))...)).....(((((.....)))))........ ( -26.60, z-score = -0.79, R) >droYak2.chrX 18079575 113 - 21770863 UAUUUGGCUUGACUUUCAUUCGUUAAGCGGUAAGUAGUUCUCAGUUCUCAAGCUUUUUUCGUUCAAGAAAAAGAUGAGCUACGGAAAUUCGGGACAAGUAGUCGCUCACAUUU ((((((.(((((.((((..(((.....)))...((((((((((((......)))((((((......)))))))).))))))).)))).))))).))))))............. ( -27.00, z-score = -1.33, R) >droSec1.super_4 4191217 94 - 6179234 AGUUUAGAUUGUCUAUG----GAUAAGUGGUAAGUGGGU---AGUCCGCAAGCUCAUCUCGUUCAAGAAAACGAUGAGCUGUGAAAAUUCGGGACAAGUUU------------ ........((((((.((----(((.........((((..---...)))).((((((..(((((......)))))))))))......)))))))))))....------------ ( -24.40, z-score = -1.79, R) >droSim1.chrX 3667262 94 + 17042790 AGUUUAGUUUGUCUAAG----GAUAAGUGGUAAGUGGAU---AGUUUUCAAGCUCAUCUUGUUCAAGAAAACGAUGAGCUGUGAAAAUUCGGGACAAGUUU------------ .((((....((((((..----.((.....))...)))))---)(((((((((((((((..(((......))))))))))).)))))))...))))......------------ ( -21.80, z-score = -1.61, R) >consensus AGUUUAGUUUGUCUUUC____GAUAAGUGGUAAGUGGAU___AGUCCUCAAGCUCAUCUCGUUCAAGAAAACGAUGAGCUGUGGAAAUUCGGGACAAGUUU____________ ......(((((((........)))))))...............((((...((((((((...((....))...)))))))).(((....))))))).................. (-14.40 = -13.48 + -0.92)

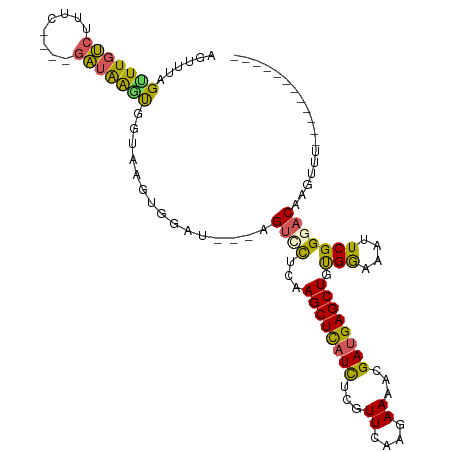
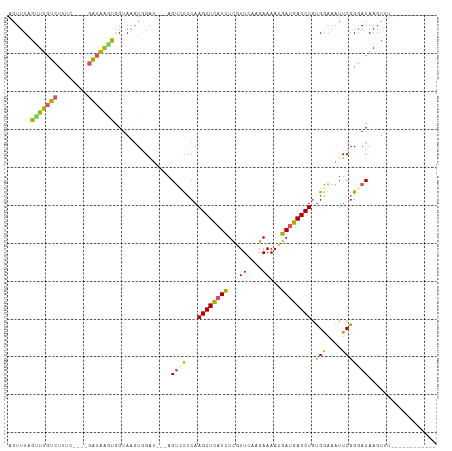
| Location | 4,755,196 – 4,755,288 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.57 |
| Shannon entropy | 0.48575 |
| G+C content | 0.39727 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -8.28 |
| Energy contribution | -8.92 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4755196 92 - 22422827 ------------AAACUUGUACCGAAUACCCACAGCUCAUCGUUUUCUUGAACGAGAUGAGCUUGAGGACU---AUCCA--UCCUAUCCUUA----GAAAGACAAACUAAACU ------------....((((.............((((((((((((....))))))..))))))((((((..---.....--.....))))))----.....))))........ ( -18.20, z-score = -2.25, R) >droEre2.scaffold_4690 2124087 110 - 18748788 AAAUGUGAGGGACUACUUGUCCCGAAUUUCCACAGCUCAUCAUUUUCUUGAACGAGAUAAGCUUGAGUGCUG---CCUACUUACCGCUUAACGUAAGGAAGUCAAACAAAAUG ...(((..(((((.....)))))((.(((((....(((.(((......)))..))).(((((.((((((...---..))))))..)))))......)))))))..)))..... ( -27.30, z-score = -1.91, R) >droYak2.chrX 18079575 113 + 21770863 AAAUGUGAGCGACUACUUGUCCCGAAUUUCCGUAGCUCAUCUUUUUCUUGAACGAAAAAAGCUUGAGAACUGAGAACUACUUACCGCUUAACGAAUGAAAGUCAAGCCAAAUA .....((((((((.....))).((......))..)))))((((.((((..(.(.......).)..))))..))))..........((((.((........)).))))...... ( -20.80, z-score = -0.94, R) >droSec1.super_4 4191217 94 + 6179234 ------------AAACUUGUCCCGAAUUUUCACAGCUCAUCGUUUUCUUGAACGAGAUGAGCUUGCGGACU---ACCCACUUACCACUUAUC----CAUAGACAAUCUAAACU ------------....(((((............((((((((((((....))))))..))))))...((...---..))..............----....)))))........ ( -18.00, z-score = -2.58, R) >droSim1.chrX 3667262 94 - 17042790 ------------AAACUUGUCCCGAAUUUUCACAGCUCAUCGUUUUCUUGAACAAGAUGAGCUUGAAAACU---AUCCACUUACCACUUAUC----CUUAGACAAACUAAACU ------------....(((((.....((((((.((((((((((((....))))..))))))))))))))..---..............((..----..)))))))........ ( -18.70, z-score = -4.14, R) >consensus ____________AAACUUGUCCCGAAUUUCCACAGCUCAUCGUUUUCUUGAACGAGAUGAGCUUGAGAACU___ACCCACUUACCACUUAUC____GAAAGACAAACUAAACU ...........................(((((.((((((((((((....))))..)))))))))).)))............................................ ( -8.28 = -8.92 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:48 2011