| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,744,557 – 4,744,659 |
| Length | 102 |
| Max. P | 0.684411 |

| Location | 4,744,557 – 4,744,659 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 57.46 |
| Shannon entropy | 0.80914 |
| G+C content | 0.45905 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -7.29 |
| Energy contribution | -5.83 |
| Covariance contribution | -1.46 |
| Combinations/Pair | 1.87 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4744557 102 - 22422827 UGAAUCAGACAGGAAAAAUGCAUCUCCUGGGUGGGU-GGGUUGUAGAA----UACCUAUAAAUAGGAUAAGUGGGUACCGUUCUACUUGUCUG--UGUCGUUGAAAAUG---- ....(((..(((((..........)))))..)))((-((((.......----.)))))).....(((((((((((......))))))))))).--..............---- ( -27.40, z-score = -1.67, R) >droSim1.chrX 3656771 110 - 17042790 UGAAUCAGACAGG-AAAAUGCAUCUCCUGGGUGGGUGGGGGAGUAGAAUAUGUACCUAUAAAUAGGAUAAGUGGGUAACGUUCUACUUGUCUG--UGUCGUUGGAAAUGCGGU ....(((..((((-(.........)))))..)))(..((.(((((((((...(((((((...........)))))))..))))))))).))..--).((((.......)))). ( -32.30, z-score = -2.40, R) >droSec1.super_4 4181093 110 + 6179234 UGAAUCAGACAGG-AAAAUGCAUCUCCUGUGUGGGUGGGAGAGUAGAAUAUGAACCUAUAAAUAGGAUAAGUGAGUAACGUUCUACAUGUCUG--UGUCGUUGGAAAUGCGGU .((..((((((..-........(((((..(....)..)))))(((((((.....((((....))))....((.....))))))))).))))))--..)).............. ( -28.90, z-score = -1.86, R) >droYak2.chrX 18068500 101 + 21770863 UGAAUCAGACAGG-AAAAUGCAACUGCCGUUUGGA--GAGGAGAA-------UGCCUAUAAAUAGGAUAAGUACGAACUCUUGUUCCUGUCUG--UGUCCUUUAAACUGAAGU .((..((((((((-((...((....))......((--(((..(.(-------(.((((....))))....)).)...))))).))))))))))--..))((((.....)))). ( -25.80, z-score = -1.86, R) >droEre2.scaffold_4690 2112866 85 - 18748788 UGAAUCAGACAGG-AAAAUGCAGCUCCUGUGUGGA--GAGGCGUA----------CUAU----------UGUAC---CUAUUGUACUUGUCUG--GGUCGUUAGAACUGCAGU .((..((((((((-.....((..((((.....)))--)..))(((----------(...----------.))))---........))))))))--..)).............. ( -28.00, z-score = -2.13, R) >dp4.chrXL_group1a 7152961 100 - 9151740 ---UGCGGAUGUGGAAACUGCAUUUGCUGGGUAGACGGUAGGCUCA-------GUAUGCCGGAGUCAAAGUUGAACGCUUUCUCCAUUGUCCGCCUGUCGUAUGGGGAGC--- ---.(((((((((....).)))))))).(((..((((((.(((...-------....)))((((..(((((.....)))))))))))))))..)))..............--- ( -34.50, z-score = -1.07, R) >droPer1.super_13 1762688 107 - 2293547 ---UGCGGAUGUGGAAACUGCAUUUGCUGGGUAGACGGCAGGCUUAGAGAAGUGUAUGCCGGAGUCAAAGUUGAACGCUUUCUCCAUUGUCCGCUUGUGGUAUGGGGAGC--- ---.(((((((((....).))))))))(((.(...(((((.((((....))))...))))).).)))............((((((((...(((....))).)))))))).--- ( -34.70, z-score = -0.90, R) >consensus UGAAUCAGACAGG_AAAAUGCAUCUCCUGGGUGGAUGGGGGAGUAG_A____UACCUAUAAAUAGGAUAAGUGAGUACUGUUCUACUUGUCUG__UGUCGUUGGAAAUGC_GU .....((((((..................(((((((.................)))))))............((........))...)))))).................... ( -7.29 = -5.83 + -1.46)

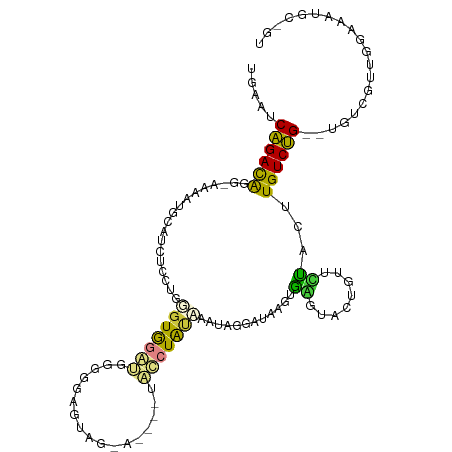
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:46 2011