
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,114,304 – 10,114,402 |
| Length | 98 |
| Max. P | 0.507298 |

| Location | 10,114,304 – 10,114,402 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Shannon entropy | 0.36447 |
| G+C content | 0.51383 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.507298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10114304 98 + 23011544 --------------------GGACUCGUAAAUGCCAUGGGCGACCAGGCCGGUUUGGCGGAAGCGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAAAAAUCU --------------------............(((.(((....))))))((((((.((....)).......))-)))).(((((((((.(........))))))))))........... ( -29.70, z-score = -1.34, R) >droGri2.scaffold_15252 3838168 86 + 17193109 --------------------UGAUUCC-AGGAGC----GGCGACAAGGUCGCUU-GCCGGAAAUGAUAACAAAAGCCGCUGACACUUGACAAAGUUGCG-AGUUAUAAAAACA------ --------------------...((((-.((...----((((((...)))))).-.))))))...........((((((....((((....)))).)))-.))).........------ ( -20.60, z-score = -0.74, R) >droSim1.chr2L 9905414 111 + 22036055 ------GGACUCU-AACUCCUGGCUCGUAAAUGCCAUGGGCGACCAGGCCGGUUUGUCGGAAGCGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAAAAAUCU ------(((....-...)))((((........))))(((....)))(((..(((.((((....)))))))...-)))..(((((((((.(........))))))))))........... ( -33.80, z-score = -1.73, R) >droSec1.super_3 5552029 110 + 7220098 ------GGACUCU-AACUCCUGGCUCUUAAAUGCCAUGGGCGACCAGGCCGGUUUGGCGGAAGCGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAAAA-UCU ------(((....-...)))((((........))))(((....)))(((..((((.((....)).).)))...-)))..(((((((((.(........)))))))))).......-... ( -34.20, z-score = -1.86, R) >droYak2.chr2L 12799184 108 + 22324452 ------CGACUCU-AACUCCUGACUCGCAAAUGCCAUGGGCGACCAGGCCGGUUUGGCGGAAGCGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAA---UCU ------.((.((.-.......)).))((....(((.(((....)))))).(((((.((....)).......))-)))))(((((((((.(........)))))))))).....---... ( -32.40, z-score = -1.46, R) >droEre2.scaffold_4929 10729484 108 + 26641161 ------UGACUCU-AACUCCUGACUCGUAAAUGCCAUGGGCGACCAGGCCGGUUUGGCGGAAGCGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAA---UCU ------.((.((.-.......)).))......(((.(((....))))))((((((.((....)).......))-)))).(((((((((.(........)))))))))).....---... ( -30.90, z-score = -1.25, R) >droAna3.scaffold_12916 13947932 106 + 16180835 ------UGACUCU-GACUUCUGACUCG-ACAAGCCCUGGGCGACCAGGUUGGUU-GGCGGAAGCGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAA---UCU ------......(-(.((((((.(.((-((....(((((....)))))...)))-))))))))))........-.....(((((((((.(........)))))))))).....---... ( -33.50, z-score = -1.85, R) >dp4.chr4_group3 11613625 113 - 11692001 CGACUGUGACUCCCGACUCCUGACUCG-ACAAGCCGUGGGCGACCAGGUUGGUU-GGCGGAAGUGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAA---UCU (((((........(((........)))-...((((.(((....)))))))))))-)((((...((....))..-.))))(((((((((.(........)))))))))).....---... ( -32.90, z-score = -0.46, R) >droPer1.super_1 8730570 113 - 10282868 CGACUGUGACUCCCGACUCCUGACUCG-ACAAGCCGUGGGCGACCAGGUUGGUU-GGCGGAAGUGAUAACAAA-GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAA---UCU (((((........(((........)))-...((((.(((....)))))))))))-)((((...((....))..-.))))(((((((((.(........)))))))))).....---... ( -32.90, z-score = -0.46, R) >consensus ______UGACUCU_AACUCCUGACUCG_AAAUGCCAUGGGCGACCAGGCCGGUUUGGCGGAAGCGAUAACAAA_GCCGCUGACACUUGACAAACUUGCGCGAGUGUCAAAAAA___UCU ................................((..(((....)))(((..(((..((....))...)))....)))))(((((((((.(........))))))))))........... (-21.39 = -21.93 + 0.55)

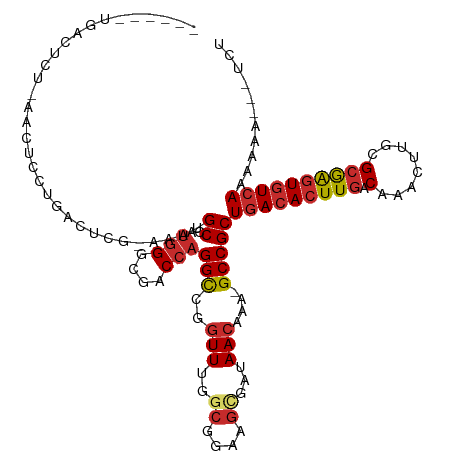

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:39 2011