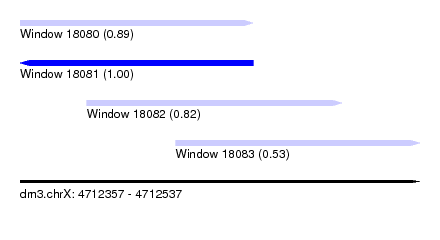
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,712,357 – 4,712,537 |
| Length | 180 |
| Max. P | 0.997439 |

| Location | 4,712,357 – 4,712,462 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 124 |
| Reading direction | forward |
| Mean pairwise identity | 71.20 |
| Shannon entropy | 0.50202 |
| G+C content | 0.37566 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
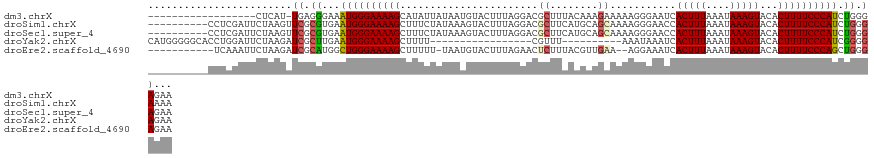
>dm3.chrX 4712357 105 + 22422827 ------------------CUCAU-UGAGGGAAAUGGGAAAAGCAUAUUAUAAUGUACUUUAGGACGCUUUACAAAGAAAAAGGGAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAA ------------------(((..-...((...((((((((((..........(((((((((.....(((....)))...((((......))))...)))))))))))))))))))))..))).. ( -22.50, z-score = -1.81, R) >droSim1.chrX 3635370 114 + 17042790 ----------CCUCGAUUCUAAGUUCGCGUGAAUGGGAAAAGCUUUCUAUAAAGUACUUUAGGACGCUUCAUGCAGCAAAAGGGAACCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAAAA ----------.............(((.((.((.(((((((((.(((....)))((((((((....(((......))).(((((....).))))...)))))))).))))))))))))).))).. ( -25.90, z-score = -0.99, R) >droSec1.super_4 4156896 114 - 6179234 ----------CCUCGAUUCUAAGUUCGCGUGAAUGGGAAAAGCUUUCUAUAAAGUACUUUAGGACGCUUCAUGCAGCAAAAGGGAACCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAA ----------.............(((.((.((.(((((((((.(((....)))((((((((....(((......))).(((((....).))))...)))))))).))))))))))))).))).. ( -25.60, z-score = -0.53, R) >droYak2.chrX 18043987 97 - 21770863 CAUGGGGGCACCUGGAUUCUAAGAUCGCUUGAAUGGGAAAAGCUUUU-----------------CGUUU----------AAAUAAAUCACUUUAAAUAAAGUACACUUUUCCCAUCGGGGAGAA ...(((....))).((((....)))).(((((.(((((((((..(((-----------------..(((----------(((........))))))..)))....))))))))))))))..... ( -23.00, z-score = -1.10, R) >droEre2.scaffold_4690 2088918 110 + 18748788 -----------UCAAAUUCUAAGAUCGCAUGGCUGGGAAAAGCUUUUU-UAAUGUACUUUAGAACUCUUUACGUUGAA--AGGAAAUCACUUUAAAUAAAGUACACUUUUCCCAGCUGGGAGAA -----------.............((...(((((((((((((.(((((-(((((((....((....)).)))))))))--))).....(((((....)))))...)))))))))))))...)). ( -30.90, z-score = -3.46, R) >consensus __________CCUCGAUUCUAAGAUCGCGUGAAUGGGAAAAGCUUUUUAUAAAGUACUUUAGGACGCUUCACGCAGAAAAAGGGAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAA ........................((.((...((((((((((.......................((........))...........(((((....)))))...)))))))))).)).))... (-14.14 = -14.22 + 0.08)
| Location | 4,712,357 – 4,712,462 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 71.20 |
| Shannon entropy | 0.50202 |
| G+C content | 0.37566 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -19.27 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.997439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4712357 105 - 22422827 UUCUCCCAGAUGGGAAAAGUGUACUUUAUUUAAAGUGAUUCCCUUUUUCUUUGUAAAGCGUCCUAAAGUACAUUAUAAUAUGCUUUUCCCAUUUCCCUCA-AUGAG------------------ .......((((((((((((((((((((((..((((.((.......)).)))))))))).((.(....).)).......))))))))))))))))......-.....------------------ ( -21.60, z-score = -3.13, R) >droSim1.chrX 3635370 114 - 17042790 UUUUCCCAGAUGGGAAAAGUGUACUUUAUUUAAAGUGGUUCCCUUUUGCUGCAUGAAGCGUCCUAAAGUACUUUAUAGAAAGCUUUUCCCAUUCACGCGAACUUAGAAUCGAGG---------- .....((.((((((((((((...((.....(((((((.((......((((......)))).....)).))))))).))...))))))))))))....(((........))).))---------- ( -27.70, z-score = -1.59, R) >droSec1.super_4 4156896 114 + 6179234 UUCUCCCAGAUGGGAAAAGUGUACUUUAUUUAAAGUGGUUCCCUUUUGCUGCAUGAAGCGUCCUAAAGUACUUUAUAGAAAGCUUUUCCCAUUCACGCGAACUUAGAAUCGAGG---------- .....((.((((((((((((...((.....(((((((.((......((((......)))).....)).))))))).))...))))))))))))....(((........))).))---------- ( -27.70, z-score = -1.46, R) >droYak2.chrX 18043987 97 + 21770863 UUCUCCCCGAUGGGAAAAGUGUACUUUAUUUAAAGUGAUUUAUUU----------AAACG-----------------AAAAGCUUUUCCCAUUCAAGCGAUCUUAGAAUCCAGGUGCCCCCAUG ........((((((((((((....((((..((((....))))..)----------)))..-----------------....))))))))))))...((.((((........))))))....... ( -21.70, z-score = -2.32, R) >droEre2.scaffold_4690 2088918 110 - 18748788 UUCUCCCAGCUGGGAAAAGUGUACUUUAUUUAAAGUGAUUUCCU--UUCAACGUAAAGAGUUCUAAAGUACAUUA-AAAAAGCUUUUCCCAGCCAUGCGAUCUUAGAAUUUGA----------- ........((((((((((((((((((((((((...(((......--.)))...))))......))))))))....-.....))))))))))))....................----------- ( -26.70, z-score = -2.90, R) >consensus UUCUCCCAGAUGGGAAAAGUGUACUUUAUUUAAAGUGAUUCCCUUUUGCUGCAUAAAGCGUCCUAAAGUACAUUAUAAAAAGCUUUUCCCAUUCACGCGAACUUAGAAUCGAGG__________ ........((((((((((((.((((((....)))))).....((((........)))).......................))))))))))))............................... (-19.27 = -18.84 + -0.43)
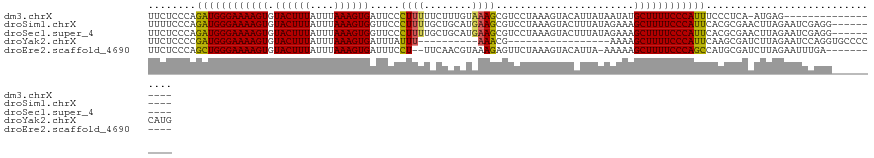
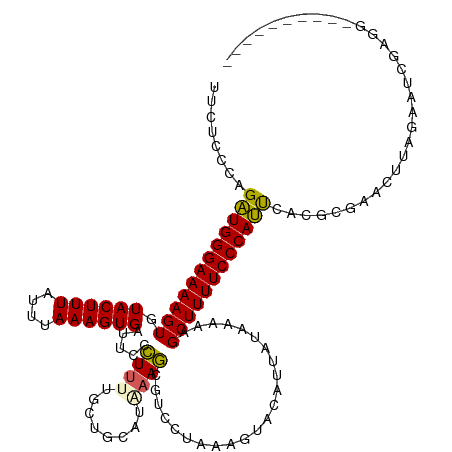
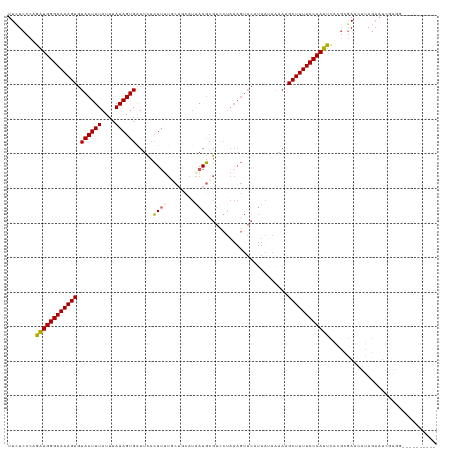
| Location | 4,712,387 – 4,712,502 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Shannon entropy | 0.25163 |
| G+C content | 0.33674 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.96 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4712387 115 + 22422827 UAAUGUACUUUAGGACGCUUUACAAAGAAAAAGGGAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCAAAUUUGCGUUAAAAUAAACAGCAAGAAA ...(((..((((.(((((...........(((((((...(((((....)))))....(((((((....))))))))))))))..........)))))....))))..)))..... ( -24.10, z-score = -2.22, R) >droSim1.chrX 3635409 115 + 17042790 UAAAGUACUUUAGGACGCUUCAUGCAGCAAAAGGGAACCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAAAAUCCUUUUCAGCGAAUUUGUGUUAAAAUAAACAGCAAGAAA ..................(((.(((.((.(((((((...(((((....)))))....(((((((....))))))))))))))..)).......((((......))))))).))). ( -24.40, z-score = -1.63, R) >droSec1.super_4 4156935 115 - 6179234 UAAAGUACUUUAGGACGCUUCAUGCAGCAAAAGGGAACCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGUGUUAAAAUAAACAGCAAGAAA ..................(((.(((.((.(((((((...(((((....)))))....(((((((....))))))))))))))..)).......((((......))))))).))). ( -24.50, z-score = -1.40, R) >droYak2.chrX 18044018 106 - 21770863 ------AAUGGGAAAAGCUUUUCGUUU---AAAUAAAUCACUUUAAAUAAAGUACACUUUUCCCAUCGGGGAGAAUCCCUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAA ------.((((((((((..(((..(((---(((........))))))..)))....)))))))))).((((.....)))).........((((((((......))).)))))... ( -24.60, z-score = -2.60, R) >droEre2.scaffold_4690 2088955 113 + 18748788 UAAUGUACUUUAGAACUCUUUACGUUG--AAAGGAAAUCACUUUAAAUAAAGUACACUUUUCCCAGCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAA ......................(((((--((((((....(((((....)))))....(((((((....)))))))))).))))))))..((((((((......))).)))))... ( -27.50, z-score = -3.09, R) >consensus UAAAGUACUUUAGGACGCUUUACGCAG_AAAAGGGAAUCACUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAA ......................(((...(((((((....(((((....)))))....(((((((....))))))))))))))..)))..((((((((......))).)))))... (-17.12 = -17.96 + 0.84)

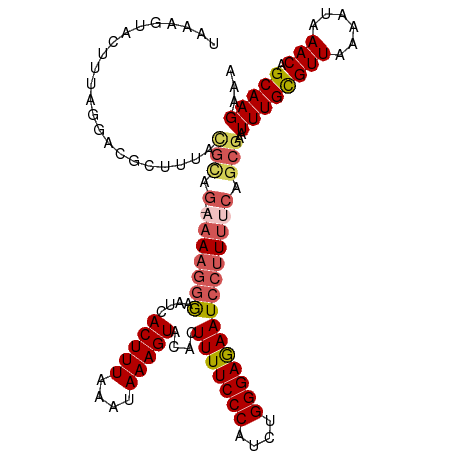
| Location | 4,712,427 – 4,712,537 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.59 |
| Shannon entropy | 0.16307 |
| G+C content | 0.33270 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -17.04 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
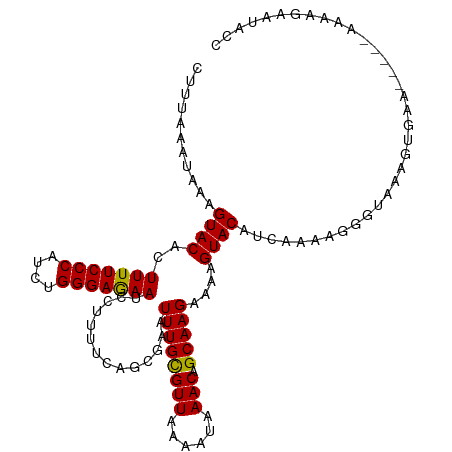
>dm3.chrX 4712427 110 + 22422827 CUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCAAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAGUGAA-----CAAAGAAUACC ...........((.((((((.(((..(((..((.....))..)))....((((((((......))).)))))...............))).)))))).)-----).......... ( -22.10, z-score = -2.37, R) >droSim1.chrX 3635449 110 + 17042790 CUUUAAAUAAAGUACACUUUUCCCAUCUGGGAAAAUCCUUUUCAGCGAAUUUGUGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAGUGAA-----AAAAGAAUACC ..............((((((((((....))))).((((((((..((...(((((......)))))..))..((........)))))))))).)))))..-----........... ( -20.20, z-score = -1.77, R) >droSec1.super_4 4156975 110 - 6179234 CUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGUGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAGUGAA-----AAAAGAAUACC ..............((((((((((....))))))((((((((..((...(((((......)))))..))..((........))))))))))..))))..-----........... ( -20.30, z-score = -1.55, R) >droYak2.chrX 18044049 115 - 21770863 CUUUAAAUAAAGUACACUUUUCCCAUCGGGGAGAAUCCCUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAAAAAAAAUGGGGAAAAACACC .................(((((((((..........((((((..((...((((((((......))).)))))....))......))))))..........)))))))))...... ( -20.35, z-score = -0.87, R) >droEre2.scaffold_4690 2088993 108 + 18748788 CUUUAAAUAAAGUACACUUUUCCCAGCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACGUCAAAAGGGUAAAA-------GGGAAAAAACACC .................(((((((.((((..((.....))..))))...((((((((......))).))))).......................-------)))))))...... ( -22.30, z-score = -1.58, R) >consensus CUUUAAAUAAAGUACACUUUUCCCAUCUGGGAGAAUCCUUUUCAGCGAAUUUGCGUUAAAAUAAACAGCAAGAAAAGUACAUCAAAAGGGUAAAGUGAA_____AAAAGAAUACC ...........((((..(((((((....)))))))..............((((((((......))).)))))....))))................................... (-17.04 = -16.64 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:41 2011