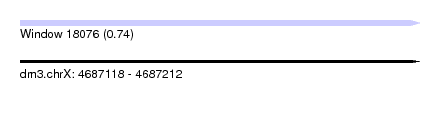
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,687,118 – 4,687,212 |
| Length | 94 |
| Max. P | 0.740043 |

| Location | 4,687,118 – 4,687,212 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.53 |
| Shannon entropy | 0.26471 |
| G+C content | 0.31480 |
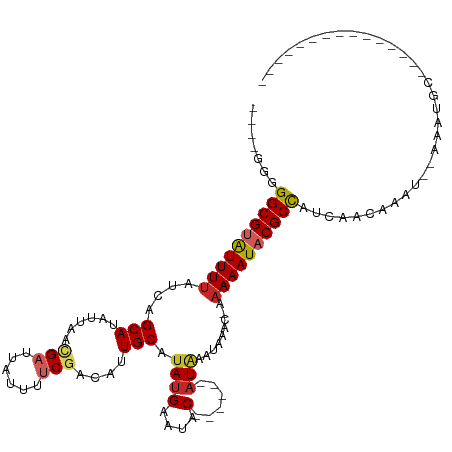
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -12.11 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740043 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
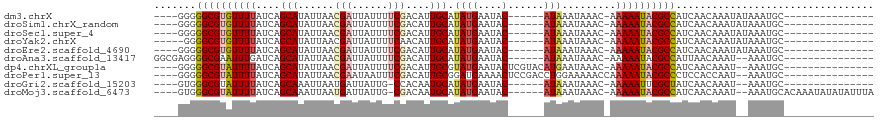
>dm3.chrX 4687118 94 + 22422827 ----GGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUACGCCAUCAACAAAUAUAAAUGC--------------- ----(..((((((((((....(((......(((......)))....))).((((....)------))).......-.))))))))))..)...............--------------- ( -19.60, z-score = -2.84, R) >droSim1.chrX_random 1663991 94 + 5698898 ----GGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUACGCCAUCAACAAAUAUAAAUGC--------------- ----(..((((((((((....(((......(((......)))....))).((((....)------))).......-.))))))))))..)...............--------------- ( -19.60, z-score = -2.84, R) >droSec1.super_4 4129352 94 - 6179234 ----GGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUACGCCAUCAACAAAUAUAAAUGC--------------- ----(..((((((((((....(((......(((......)))....))).((((....)------))).......-.))))))))))..)...............--------------- ( -19.60, z-score = -2.84, R) >droYak2.chrX 18012881 93 - 21770863 -----GGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCAACAUUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUACGCCAUCAACAAAUAUAAAUGC--------------- -----..((((((((((....(((.......((......)).....))).((((....)------))).......-.))))))))))..................--------------- ( -16.40, z-score = -1.92, R) >droEre2.scaffold_4690 2060597 94 + 18748788 ----GGGGGCGUGUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUACGCCAUCAACAAAUAUAAAUGC--------------- ----(..((((((((((....(((......(((......)))....))).((((....)------))).......-.))))))))))..)...............--------------- ( -19.60, z-score = -2.84, R) >droAna3.scaffold_13417 6952439 96 - 6960332 GGCGAGGGGCGAAUUUGAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUACGCCAUUAACAAAU--AAAUGC--------------- ((((.(...((((..(((((...........)))))..)))).)......((((....)------))).......-.......))))..........--......--------------- ( -15.60, z-score = -1.36, R) >dp4.chrXL_group1a 2063811 98 - 9151740 ----GGGGGCGUAUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCGUAUGAAUACUCGUACAUGAAUAAAC-AAAAAUACGCCAUCAACAAAU--AAAUGC--------------- ----(..((((((((((....(((......(((......)))....)))((((((....))))))..........-.))))))))))..).......--......--------------- ( -24.60, z-score = -3.78, R) >droPer1.super_13 598969 99 - 2293547 ----GGGGGCGUAUUUUAUCAGCAUAUUAACGAAUAAUUUCGACAUUGCGGAUGAAAACUCCGACCUGGAAAAACCAAAAAUACGCCCUCCACCAAU--AAAUGC--------------- ----(((((((((((((.((.(((......((((....))))....)))((((....).)))))..(((.....)))))))))))))).))......--......--------------- ( -28.90, z-score = -4.04, R) >droGri2.scaffold_15203 2104746 91 - 11997470 ----GUGGGCGUAUUUUAUCAGCAAAUUAAUGAUUAUUG-CCACAAUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUUCGCUAUCAACAAAU--AAAUGC--------------- ----((.((((.(((((....................((-(......)))((((....)------))).......-.))))).))))....))....--......--------------- ( -9.70, z-score = 1.37, R) >droMoj3.scaffold_6473 8579670 106 + 16943266 ----GUGGGCGUAUUUUAUCAGCAAAUUAAUGAUUAUUG-CGACAAUGCAUAUGAAUAC------AUAAAUAAAC-AAAAAUACGCCAUCAACAAAU--AAAUGCACAAAUAUAUAUUUA ----(((((((((((((....((((...........)))-).........((((....)------))).......-.)))))))))).....((...--...)))))............. ( -17.40, z-score = -0.87, R) >consensus ____GGGGGCGUAUUUUAUCAGCAUAUUAACGAUUAUUUUCGACAUUGCAUAUGAAUAC______AUAAAUAAAC_AAAAAUACGCCAUCAACAAAU__AAAUGC_______________ .......((((((((((.............(((......))).(((.....))).......................))))))))))................................. (-12.11 = -12.60 + 0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:35 2011