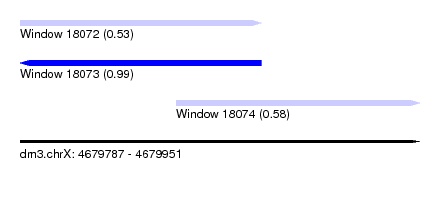
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,679,787 – 4,679,951 |
| Length | 164 |
| Max. P | 0.990284 |

| Location | 4,679,787 – 4,679,886 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.97 |
| Shannon entropy | 0.12040 |
| G+C content | 0.46506 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -25.30 |
| Energy contribution | -26.34 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
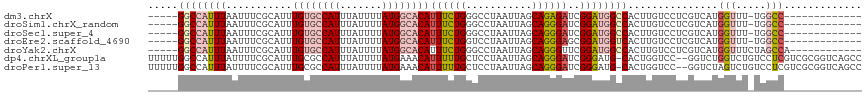
>dm3.chrX 4679787 99 + 22422827 AAAACCGA---CGAACGGCGGCCAUCAGGACAUCA--------AUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUA ......((---((....((((((((...((.....--------.))..)))))))).))))...((((((....)))))).........(((....)))........... ( -30.10, z-score = -1.36, R) >droSim1.chrX_random 1657230 102 + 5698898 GAAACUAAAACCGAACGGCGGCCAUCAGGACAUCA--------AUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUA ((((.((((...((...((((((((...((.....--------.))..))))))))...))...((((((....)))))))))).))))(((....)))........... ( -32.20, z-score = -2.26, R) >droSec1.super_4 4122732 102 - 6179234 GAAACUAAAACCGAACGGCGGCCAUCAGGACAUCA--------AUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUA ((((.((((...((...((((((((...((.....--------.))..))))))))...))...((((((....)))))))))).))))(((....)))........... ( -32.20, z-score = -2.26, R) >droEre2.scaffold_4690 2053849 101 + 18748788 GAAACUAAAACCGAACGGCGGCCAUCGGGACAUCA--------AUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGG-CCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUA ((((.((((...((...((((((((...((.....--------.))..))))))))...))...(((((.....))-))))))).))))(((....)))........... ( -30.30, z-score = -1.59, R) >droYak2.chrX 18005240 110 - 21770863 GAAACUAAAACCGAACGACGGCCAUCGGGACAUCAGGACAUCAAUCAAAUGGCCCCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUA ((((.((((...(..(((((((((..(((((((..((.(((.......))).))..))))))).)))))..))))..)..)))).))))(((....)))........... ( -32.60, z-score = -1.72, R) >consensus GAAACUAAAACCGAACGGCGGCCAUCAGGACAUCA________AUCAAAUGGCCGCAUGUCCUUUGGCCUGGUCGGGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUA ((((.((((...((...((((((((...((..............))..))))))))...))...((((((....)))))))))).))))(((....)))........... (-25.30 = -26.34 + 1.04)

| Location | 4,679,787 – 4,679,886 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Shannon entropy | 0.12040 |
| G+C content | 0.46506 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4679787 99 - 22422827 UAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAU--------UGAUGUCCUGAUGGCCGCCGUUCG---UCGGUUUU .........((((.................(((((......)))))..((((..(((((((((.(((--------....)))..))))))))).)))))---)))..... ( -33.60, z-score = -2.13, R) >droSim1.chrX_random 1657230 102 - 5698898 UAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAU--------UGAUGUCCUGAUGGCCGCCGUUCGGUUUUAGUUUC ...........(((....)))((((((((((((((......)))))..((((..(((((((((.(((--------....)))..))))))))).))))...))))))))) ( -37.20, z-score = -3.50, R) >droSec1.super_4 4122732 102 + 6179234 UAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAU--------UGAUGUCCUGAUGGCCGCCGUUCGGUUUUAGUUUC ...........(((....)))((((((((((((((......)))))..((((..(((((((((.(((--------....)))..))))))))).))))...))))))))) ( -37.20, z-score = -3.50, R) >droEre2.scaffold_4690 2053849 101 - 18748788 UAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGG-CCGACCAGGCCAAAGGACAUGCGGCCAUUUGAU--------UGAUGUCCCGAUGGCCGCCGUUCGGUUUUAGUUUC ...........(((....)))((((((((((((-((.....)))))..((((..(((((((((.(((--------....)))..))))))))).))))...))))))))) ( -37.30, z-score = -3.47, R) >droYak2.chrX 18005240 110 + 21770863 UAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGGGGCCAUUUGAUUGAUGUCCUGAUGUCCCGAUGGCCGUCGUUCGGUUUUAGUUUC ...........(((....)))(((((((((..(((.(((..(((((..((((((.(((.((((.....)))).))).))))))...)))))...))).)))))))))))) ( -39.80, z-score = -3.12, R) >consensus UAAAAUAAAUGGCACAAAUGCGAAAUUAAAUGGCCCGACCAGGCCAAAGGACAUGCGGCCAUUUGAU________UGAUGUCCUGAUGGCCGCCGUUCGGUUUUAGUUUC ...........(((....)))((((((((((((((......)))))..((((..(((((((((.(((............)))..))))))))).))))...))))))))) (-31.00 = -31.32 + 0.32)

| Location | 4,679,851 – 4,679,951 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Shannon entropy | 0.32837 |
| G+C content | 0.45620 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -17.45 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4679851 100 + 22422827 -----GGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGAGAUCGGAUGGCCACUUGUCCUCGUCAUGGUUU-UGGCC------------- -----(((((((..(((((.....((((((((.......)))))))).......((.......)).)))))..)))))))..........((((......-)))).------------- ( -34.00, z-score = -3.05, R) >droSim1.chrX_random 1657297 100 + 5698898 -----GGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGAUCGGAUGGCCACUUGUCCUCGUCAUGGUUU-UGGCC------------- -----(((((((..((..(.....((((((((.......)))))))).......((.......)).)..))..)))))))..........((((......-)))).------------- ( -33.20, z-score = -2.43, R) >droSec1.super_4 4122799 100 - 6179234 -----GGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGAUCGGAUGGCCACUUGUCCUCGUCAUGGUUU-UGGCC------------- -----(((((((..((..(.....((((((((.......)))))))).......((.......)).)..))..)))))))..........((((......-)))).------------- ( -33.20, z-score = -2.43, R) >droEre2.scaffold_4690 2053915 100 + 18748788 -----GGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGUCCUAAUUAGCAGGGAGCGGAUGGUCACUUGUCCUCGUCAUGGUUU-UGGCC------------- -----(((((...((((..((...((((((((.......))))))))..((((.((((........)))).)))).((.(....).))..))...)))).-)))))------------- ( -31.50, z-score = -2.47, R) >droYak2.chrX 18005315 102 - 21770863 -----GGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGUUCGGAUGGCCACUUGUCCUCGUCAUGGUUUCUAGCCA------------ -----(((((..............((((((((.......))))))))..(((((((((........))))))))))))))..............((((.....))))------------ ( -34.80, z-score = -3.32, R) >dp4.chrXL_group1a 2056134 116 - 9151740 UUUUUGGCCAUUUAUUUUCGCAUUUGCGCCAUUUAUUUUAUGAAACAUUUUUGCUCCUAAUUAGCAGGGAUCGGGAUG-CACUGGUCC--GGUCUGGUCUGUCCUCGUCGCGGUCAGCC .....((((.........(((....)))......................(((((.......)))))(((((((....-..)))))))--)))).((.(((.((.......)).))))) ( -28.20, z-score = 0.09, R) >droPer1.super_13 591317 116 - 2293547 UUUUUGGCCAUUUAUUUUCGCAUUUGCGCCAUUUAUUUUAUGAAACAUUUUUGCUCCUAAUUAGCAGGGAUCGGGAUG-CACUGGUCC--GGUCUAGUCUGUCCUCGUCGCGGUCAGCC ...((((((..........((....))((....................((((((.......))))))(((.((((((-.(((((...--...))))).)))))).))))))))))).. ( -28.90, z-score = -0.45, R) >consensus _____GGCCAUUUAAUUUCGCAUUUGUGCCAUUUAUUUUAUGGCACAUUUCUGGGCCUAAUUAGCAGGGAUCGGAUGGCCACUUGUCCUCGUCAUGGUUU_UGGCC_____________ .....((((((((...........((((((((.......))))))))((((((...........))))))..))))))))....................................... (-17.45 = -18.70 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:34 2011