
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,635,275 – 4,635,368 |
| Length | 93 |
| Max. P | 0.956193 |

| Location | 4,635,275 – 4,635,368 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.07 |
| Shannon entropy | 0.40134 |
| G+C content | 0.41512 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
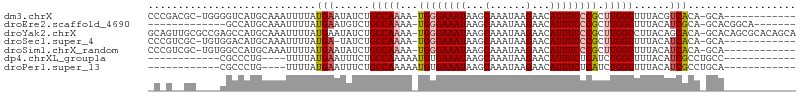
>dm3.chrX 4635275 93 + 22422827 CCCGACGC-UGGGGUCAUGCAAAUUUUAUGAAUAUCUGCCAAAA-UGGGAAAUAAGCAAAUAAGAACAUUUCCCGCUUGGCUUUACGUCACA-GCA------------ ......((-((((.(((((.......)))))...)).(((((..-((((((((...(......)...)))))))).))))).........))-)).------------ ( -23.40, z-score = -1.16, R) >droEre2.scaffold_4690 2010752 86 + 18748788 -------------GCCAUGCAAAUUUUAUGAAUGUCUGCCAAAA-UGGGAAAUAAGCAAAUAAGAACAUUUCCCGCUUGGCUUUACAUCGCA-GCACGGCA------- -------------(((.(((........((.((((..(((((..-((((((((...(......)...)))))))).)))))...))))..))-))).))).------- ( -24.10, z-score = -1.96, R) >droYak2.chrX 17959709 106 - 21770863 GCAGUUGCGCCGAGCCAUGCAAAUUUUAUGAAUAUCUGCCAAAA-UGGGAAAUAAGCAAAUAAGAACAUUUCCCGCUUGGCCUUACAGCACA-GCACAGCGCACAGCA ((.((.((((.(.((..(((.........(.....).(((((..-((((((((...(......)...)))))))).)))))......)))..-)).).)))))).)). ( -29.60, z-score = -1.52, R) >droSec1.super_4 4077644 92 - 6179234 CCCGUCGC-UGUGGACAUGCAAAUUUUAUGA-UAUCUGCCAAAA-UGGGAAAUAAGCAAAUAAGAACAUUUCCCGCUUGGCUUUACAUCACA-GCA------------ ......((-(((((.((((.......)))).-.....(((((..-((((((((...(......)...)))))))).)))))......)))))-)).------------ ( -26.40, z-score = -2.83, R) >droSim1.chrX_random 1653090 93 + 5698898 CCCGUCGC-UGUGGCCAUGCAAAUUUUAUGAAUAUCUGCCAAAA-UGGGAAAUAAGCAAAUAAGAACAUUUCCCGCUUGGCUUUACAUCACA-GCA------------ ......((-(((((.((((.......)))).......(((((..-((((((((...(......)...)))))))).)))))......)))))-)).------------ ( -25.90, z-score = -2.32, R) >dp4.chrXL_group1a 2099378 80 - 9151740 ------------CGCCCUG----UUUUAUGAAUUUCUGCCAAAAAUGUGAAAUAAGCAAAUAAGAACAUUUCUCAUCUGGCUUUACAUCGCCUGCC------------ ------------.((..((----(.............((((...(((.(((((...(......)...))))).))).))))...)))..)).....------------ ( -9.59, z-score = 0.51, R) >droPer1.super_13 635376 80 - 2293547 ------------CGCCCUG----UUUUAUGAAUUUCUGCCAAAAAUGUGAAAUAAGCAAAUAAGAACAUUUCUCAUCUGGCUUUACAUCGCCUGCA------------ ------------......(----(....(((......((((...(((.(((((...(......)...))))).))).))))......)))...)).------------ ( -9.90, z-score = 0.61, R) >consensus _C_G__GC____GGCCAUGCAAAUUUUAUGAAUAUCUGCCAAAA_UGGGAAAUAAGCAAAUAAGAACAUUUCCCGCUUGGCUUUACAUCACA_GCA____________ ............................(((......(((((...((((((((...(......)...)))))))).)))))......))).................. (-13.53 = -13.80 + 0.27)

| Location | 4,635,275 – 4,635,368 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.07 |
| Shannon entropy | 0.40134 |
| G+C content | 0.41512 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.99 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
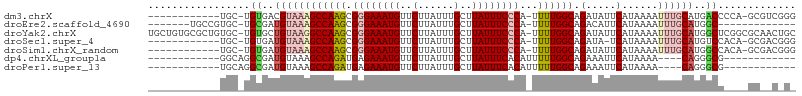
>dm3.chrX 4635275 93 - 22422827 ------------UGC-UGUGACGUAAAGCCAAGCGGGAAAUGUUCUUAUUUGCUUAUUUCCCA-UUUUGGCAGAUAUUCAUAAAAUUUGCAUGACCCCA-GCGUCGGG ------------...-..((((((...((((((.((((((((..(......)..)))))))).-.))))))......((((.........)))).....-)))))).. ( -24.90, z-score = -1.33, R) >droEre2.scaffold_4690 2010752 86 - 18748788 -------UGCCGUGC-UGCGAUGUAAAGCCAAGCGGGAAAUGUUCUUAUUUGCUUAUUUCCCA-UUUUGGCAGACAUUCAUAAAAUUUGCAUGGC------------- -------.(((((((-((.(((((...((((((.((((((((..(......)..)))))))).-.))))))..)))))))........)))))))------------- ( -30.40, z-score = -3.37, R) >droYak2.chrX 17959709 106 + 21770863 UGCUGUGCGCUGUGC-UGUGCUGUAAGGCCAAGCGGGAAAUGUUCUUAUUUGCUUAUUUCCCA-UUUUGGCAGAUAUUCAUAAAAUUUGCAUGGCUCGGCGCAACUGC .((..(((((((.((-(((((......((((((.((((((((..(......)..)))))))).-.)))))).(.....).........))))))).)))))))...)) ( -42.10, z-score = -3.73, R) >droSec1.super_4 4077644 92 + 6179234 ------------UGC-UGUGAUGUAAAGCCAAGCGGGAAAUGUUCUUAUUUGCUUAUUUCCCA-UUUUGGCAGAUA-UCAUAAAAUUUGCAUGUCCACA-GCGACGGG ------------.((-((((.......((((((.((((((((..(......)..)))))))).-.)))))).((((-(((.......)).)))))))))-))...... ( -31.10, z-score = -3.42, R) >droSim1.chrX_random 1653090 93 - 5698898 ------------UGC-UGUGAUGUAAAGCCAAGCGGGAAAUGUUCUUAUUUGCUUAUUUCCCA-UUUUGGCAGAUAUUCAUAAAAUUUGCAUGGCCACA-GCGACGGG ------------.((-(((((((((((((((((.((((((((..(......)..)))))))).-.)))))).(.....)......)))))))...))))-))...... ( -30.70, z-score = -2.82, R) >dp4.chrXL_group1a 2099378 80 + 9151740 ------------GGCAGGCGAUGUAAAGCCAGAUGAGAAAUGUUCUUAUUUGCUUAUUUCACAUUUUUGGCAGAAAUUCAUAAAA----CAGGGCG------------ ------------.....((..(((...((((((((.((((((..(......)..)))))).))))..)))).(.....).....)----))..)).------------ ( -16.30, z-score = -0.53, R) >droPer1.super_13 635376 80 + 2293547 ------------UGCAGGCGAUGUAAAGCCAGAUGAGAAAUGUUCUUAUUUGCUUAUUUCACAUUUUUGGCAGAAAUUCAUAAAA----CAGGGCG------------ ------------.....((..(((...((((((((.((((((..(......)..)))))).))))..)))).(.....).....)----))..)).------------ ( -16.30, z-score = -0.51, R) >consensus ____________UGC_UGUGAUGUAAAGCCAAGCGGGAAAUGUUCUUAUUUGCUUAUUUCCCA_UUUUGGCAGAUAUUCAUAAAAUUUGCAUGGCC____GC__C_G_ .................((..((((((((((((.((((((((..(......)..))))))))...)))))).(.....)......))))))..))............. (-16.04 = -16.99 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:23 2011