| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,627,380 – 4,627,430 |
| Length | 50 |
| Max. P | 0.794274 |

| Location | 4,627,380 – 4,627,430 |
|---|---|
| Length | 50 |
| Sequences | 4 |
| Columns | 50 |
| Reading direction | forward |
| Mean pairwise identity | 57.00 |
| Shannon entropy | 0.72961 |
| G+C content | 0.37000 |
| Mean single sequence MFE | -8.95 |
| Consensus MFE | -4.19 |
| Energy contribution | -4.00 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
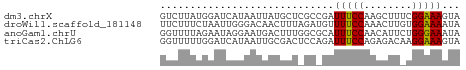
>dm3.chrX 4627380 50 + 22422827 GUCUUAUGGAUCAUAAUUAUGCUCGCCGAUUUCCAAGCUUUCGGAAAGUA ...................((((..((((...........))))..)))) ( -7.70, z-score = -0.09, R) >droWil1.scaffold_181148 3777805 50 + 5435427 UUCUUUCUAAUUGGGACAACUUUAGAUGUUUUCCAAACUUGUGGAAAAUA .....(((((..((.....)))))))(((((((((......))))))))) ( -9.70, z-score = -1.48, R) >anoGam1.chrU 3206203 50 - 59568033 GGUUUUAGAAUAGGAAUGACUUUGGCGCAUUUCCAACAUUCUGGGAAAUA ..(((((((((.((((((.(......)))))))....))))))))).... ( -8.80, z-score = -0.66, R) >triCas2.ChLG6 13069986 50 + 13544221 GGUUUUUGGAUCAUAAUUGCGACUCCAGAUUUCCAGAGACAAGGAAAGUA ....((((((((........)).))))))(((((........)))))... ( -9.60, z-score = -0.68, R) >consensus GGCUUUUGAAUCAGAAUUACGUUAGCAGAUUUCCAAACUUAUGGAAAAUA .............................(((((........)))))... ( -4.19 = -4.00 + -0.19)

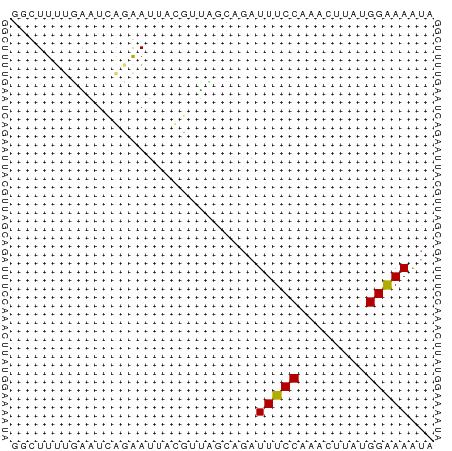
| Location | 4,627,380 – 4,627,430 |
|---|---|
| Length | 50 |
| Sequences | 4 |
| Columns | 50 |
| Reading direction | reverse |
| Mean pairwise identity | 57.00 |
| Shannon entropy | 0.72961 |
| G+C content | 0.37000 |
| Mean single sequence MFE | -7.47 |
| Consensus MFE | -2.80 |
| Energy contribution | -3.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
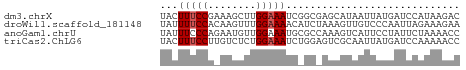
>dm3.chrX 4627380 50 - 22422827 UACUUUCCGAAAGCUUGGAAAUCGGCGAGCAUAAUUAUGAUCCAUAAGAC ...(((((((....)))))))..((....(((....)))..))....... ( -6.80, z-score = -0.16, R) >droWil1.scaffold_181148 3777805 50 - 5435427 UAUUUUCCACAAGUUUGGAAAACAUCUAAAGUUGUCCCAAUUAGAAAGAA ........((((.((((((.....)))))).))))............... ( -7.50, z-score = -1.33, R) >anoGam1.chrU 3206203 50 + 59568033 UAUUUCCCAGAAUGUUGGAAAUGCGCCAAAGUCAUUCCUAUUCUAAAACC ........((((((..((((..((......))..))))))))))...... ( -7.50, z-score = -1.61, R) >triCas2.ChLG6 13069986 50 - 13544221 UACUUUCCUUGUCUCUGGAAAUCUGGAGUCGCAAUUAUGAUCCAAAAACC ...(((((........)))))..((((.(((......)))))))...... ( -8.10, z-score = -0.97, R) >consensus UACUUUCCAAAAGUUUGGAAAUCAGCAAAAGUAAUUACGAUCCAAAAAAC ...(((((........)))))............................. ( -2.80 = -3.05 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:21 2011