| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,598,187 – 4,598,303 |
| Length | 116 |
| Max. P | 0.657697 |

| Location | 4,598,187 – 4,598,303 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.74661 |
| G+C content | 0.51719 |
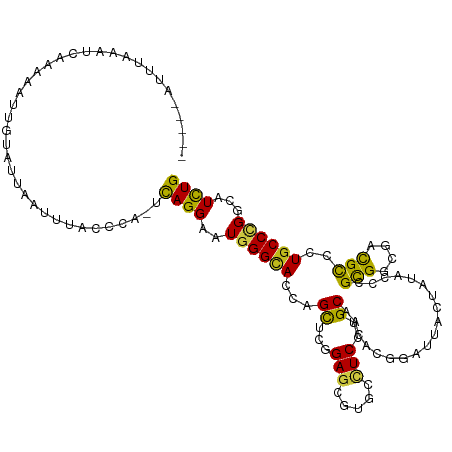
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -14.76 |
| Energy contribution | -13.70 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.31 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
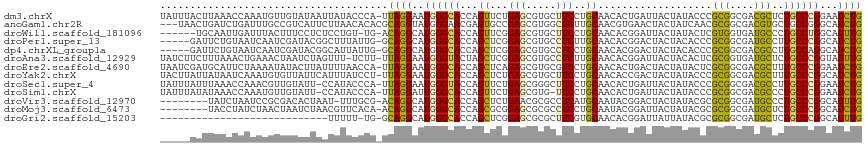
>dm3.chrX 4598187 116 + 22422827 UAUUUACUUAAACCAAAUGUUGUAUAAUUAUACCCA-UUAGGAAUGGGCACCAGUUCUGAGCGUGCUUCCUGCAACACUGAUUACUAUACCCGCGGCGACGCUCUGCCCGGAAUCUG ............((.((((..((((....)))).))-)).))..((((((..(((..(.((.((((.....)).)).)).)..)))......(((....)))..))))))....... ( -26.80, z-score = -1.03, R) >anoGam1.chr2R 31472861 114 - 62725911 ---UAACUGAUCUGAUUUGCCGUCAUUCUUAACACACGCAGGUAUGGGAACGAGUGCCGAGCGUGGCUCUUGCAACGUGAACUACUAUCAACGCGGCGACGUGCUGCCGGGCAUCUG ---....((.((((....((((((..........(((((.(((((........)))))..)))))......((..(((((.......)).)))..)))))).))...)))))).... ( -31.10, z-score = 0.94, R) >droWil1.scaffold_181096 7913347 109 - 12416693 ------UGCAAUUGAUUUACUUUCCUCUCCUGU-UG-ACAGGCAUGGGCACCAGUUCGGAGCGUGCUUCCUGCAACACGGAUUACUAUACUCGUGGUGAUGCCCUGCCUGGCAUUUG ------(((.............((..(....).-.)-)((((((.(((((.((((..((((....))))..))..((((..(......)..)))).)).)))))))))))))).... ( -33.10, z-score = -1.18, R) >droPer1.super_13 1617704 111 + 2293547 -----GAUUCUGUAAUCAAUCGAUACGGCUUUAUUG-GCAGGCAUGGGCACCAGCUCGGAGCGUGCCUCUUGCAACACGGACUACUACACCCGCGGCGAUGCCUUGCCCGGCAUCUG -----((..(((((.((....)))))))..))...(-(((.((.(((((....)))))..)).))))....((....(((..........)))..))((((((......)))))).. ( -33.90, z-score = -0.06, R) >dp4.chrXL_group1a 7013121 111 + 9151740 -----GAUUCUGUAAUCAAUCGAUACGGCAUUAUUG-GCAGGCAUGGGCACCAGCUCGGAGCGUGCCUCCUGCAACACGGACUACUACACCCGCGGCGACGCCUUGCCAGGCAUCUG -----(((((((((.((....)))))((((......-(((((...((((((..((.....)))))))))))))...................(((....)))..))))))).))).. ( -32.90, z-score = 0.33, R) >droAna3.scaffold_12929 3176933 115 + 3277472 UAUCUUCUUUAAACUGAAACUAAUCUAGUUU-UCUU-UUAGGAAUGGGUACUAGCUCGGAGCGUGCCUCUUGCAACACGGAUUACUACACUCGCGGUGAUGCUCUGCCCGGUAUUUG ....(((((.(((..(((((((...))))))-)..)-))))))).((((((..(..((((((.(((.....)))......(((((..(....)..))))))))))).)..)))))). ( -24.80, z-score = -0.01, R) >droEre2.scaffold_4690 1975099 116 + 18748788 UAAUCGAUGCAUUCUAAAAUAUACUUAUUUUAACCA-UUAGGAAUGGGCACCAGCUCAGAGCGUGCGUCCUGCAACACUGACUACUAUACUCGCGGCGACGCUUUGCCCGGAAUCUG .....((((.....(((((((....)))))))..))-)).(((.((((((...((.....))..(((((((((...................)))).)))))..))))))...))). ( -27.31, z-score = -0.96, R) >droYak2.chrX 17928406 116 - 21770863 UACUUAUUAUAAUCAAAUGUGUUAUUCAUUUAUCCU-UUAGGAAUGGGCACCAGCUCUGAGCGUGCUUCCUGCAACACCGACUACUAUACCCGCGGCGACGCUUUGCCCGGCAUCUG .............((.((((............(((.-...))).((((((........((((((((.....))..(.(((.............))).))))))))))))))))).)) ( -24.12, z-score = 0.11, R) >droSec1.super_4 4047008 115 - 6179234 UAUUUAUUUAAACCAAACGUUGUAUU-CCAUACCCA-UUAGGAAUGGGUACCAGUUCUGAGCGGGCUUCCUGCAACACUGAUUACUAUACCCGCGGCGACGCCUUGCCCGGAAUCUG ..........................-...((((((-(.....))))))).((((((((.((((((.((((((...................)))).)).))).))).))))).))) ( -29.51, z-score = -1.23, R) >droSim1.chrX 3564883 114 + 17042790 UAUUUAUAUAAACCAAAUGUUGUAUU-CCAUACCCA-UUAGGAUUGGGCACCAGUUCUGAGCGUG-UUCCUGCAACACUGAUUACUAUACCCGCGGCGACGCCCUGCCCGGAAUCUG .................(((((((..-..((((...-((((((((((...))))))))))..)))-)...))))))).............(((.((((......)))))))...... ( -28.40, z-score = -1.37, R) >droVir3.scaffold_12970 7770959 107 - 11907090 --------UAUCUAAUCCGCGACACUAAU-UUUGCG-ACAGGCAUGGGCACCAGCUCUGAACGCGCCUCAUGCAAUACGGACUACUAUACGCGCGGCGAUGCCCUGCCCGGCAUUUG --------........(((((........-..(.((-....((((((((....((.......))))).)))))....)).)..........))))).((((((......)))))).. ( -28.15, z-score = 0.27, R) >droMoj3.scaffold_6473 8236522 108 - 16943266 --------UACCUAUCUAACUAAUCUAACGUUCACA-ACAGGCAUGGGCACCAGCGCGGAGCGCGCCUCCUGCAAUACGGAUUACUAUACGCGCGGCGAUGCUCUGCCCGGCAUCUG --------............................-.((((..((((((..((((((..(((((..(((........)))........)))))..)).)))).))))))...)))) ( -28.00, z-score = 0.68, R) >droGri2.scaffold_15203 9814350 87 + 11997470 ----------------------------UUUUU-UG-GCAGGCAUGGGCACCAGCUCGGAGCGCGCUUCGUGCAACACGGAUUAUUAUACGCGCGGCGAUGCUCUGCCCGGCAUUUG ----------------------------.....-..-....((.((((((..((((((..(((((.((((((...))))))........)))))..))).))).))))))))..... ( -30.80, z-score = -0.47, R) >consensus _____AUUUAAAUCAAAAAUUGUAUUAAUUUACCCA_UCAGGAAUGGGCACCAGCUCGGAGCGUGCCUCCUGCAACACGGAUUACUAUACCCGCGGCGACGCCCUGCCCGGCAUCUG ......................................((((..((((((...((...(((.....)))..))...................(((....)))..))))))...)))) (-14.76 = -13.70 + -1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:20 2011