| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,582,070 – 4,582,154 |
| Length | 84 |
| Max. P | 0.830828 |

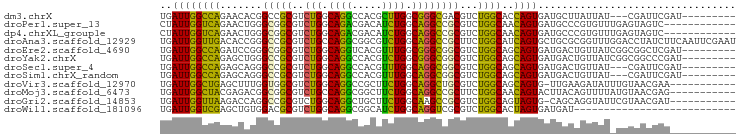
| Location | 4,582,070 – 4,582,154 |
|---|---|
| Length | 84 |
| Sequences | 12 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 68.25 |
| Shannon entropy | 0.66857 |
| G+C content | 0.60154 |
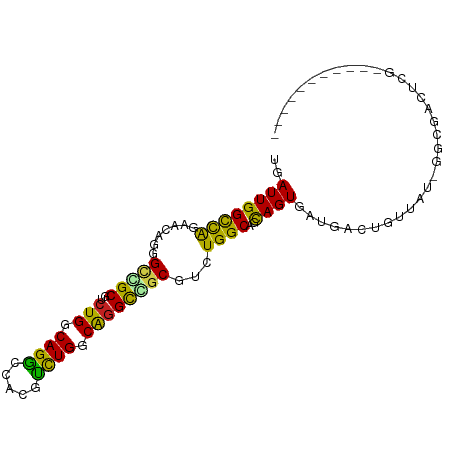
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -16.61 |
| Energy contribution | -15.40 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4582070 84 - 22422827 UGAUUGGCCAGAACACGGCCGCGUCUGGCAGGCCACGCUUGGCGGGCGACGUCUGGCACCAGUGAUGCUUAUUAU---CGAUUCGAU--------- .(((((((((((...((....((((((.(((((...))))).)))))).))))))))..((....))........---)))))....--------- ( -28.60, z-score = -0.04, R) >droPer1.super_13 888634 84 - 2293547 CUAUUGGUCAGAACUGGGCGGCGUCUGGCAGACGACAUCUGGCAGGCCGCGUCUGGCAACAGUGAUGCCCGUGUUUGAGUAGUC------------ ..((((.(((((..(((((((((((((.((((.....)))).)))).)))..(((....)))...))))))..))))).)))).------------ ( -35.50, z-score = -2.11, R) >dp4.chrXL_group1e 4855347 84 + 12523060 CUAUUGGUCAGAACUGGGCGGCGUCUGGCAGACGACAUCUGGCAGGCCGCGUCUGGCAACAGUGAUGCCCGUGUUUGAGUAGUC------------ ..((((.(((((..(((((((((((((.((((.....)))).)))).)))..(((....)))...))))))..))))).)))).------------ ( -35.50, z-score = -2.11, R) >droAna3.scaffold_12929 3158941 96 - 3277472 UGAUUGGUUGACACCGGGCCGCGUCUGCCAGGCGGCGUCUGGCAGGCCGCUUCUGGCAUCAGUGCUGCGCGGUUUGGACCUAUCUUCAAUUCGAAU .(((.((((.(.((((.((.(((((((((((((...))))))))))).((.....))......)).)).)))).).)))).)))............ ( -42.30, z-score = -2.06, R) >droEre2.scaffold_4690 1959376 87 - 18748788 UGAUUGGCCAGAUCCGGGCGGCGUCUGGCAGGUCACGUUUGGCGGGCGGCGUCUGGCAGCAGUGAUGACUGUUAUCGGCGGCUCGAU--------- .((.(.(((....(((((((.((((((.((((.....)))).)))))).))))))).((((((....))))))...))).).))...--------- ( -38.40, z-score = -1.45, R) >droYak2.chrX 17912651 87 + 21770863 UGAUUGGCCAGAGCUGGGCCGCGUCUGGCAGGCCACGUCUGGCGGGCGGCGUCUGGCAGCAGUGAUGACUGUUAUCGGCGGCCCGAU--------- ......((((((....((((((.....)).))))...))))))((((.((...((((((((....)).))))))...)).))))...--------- ( -41.70, z-score = -0.95, R) >droSec1.super_4 4031412 84 + 6179234 UGAUUGGCCAGAGCAGGGCCGCGUCUGGCAGGCCACGUUUGGCAGGCGGCGUCUGGCAGCAGUGAUGACUGUUAU---CGAUUCGAU--------- .(((((((((((.....(((((..(((.(((((...))))).)))))))).))))))((((((....))))))..---)))))....--------- ( -33.10, z-score = -0.78, R) >droSim1.chrX_random 1625439 84 - 5698898 UGAUUGGCCAGAGCAGGGCCGCGUCUGGCAGGCCACGUUUGGCAGGCGGCGUCUGGCAGCAGUGAUGACUGUUAU---CGAUUCGAU--------- .(((((((((((.....(((((..(((.(((((...))))).)))))))).))))))((((((....))))))..---)))))....--------- ( -33.10, z-score = -0.78, R) >droVir3.scaffold_12970 7750585 84 + 11907090 UGAUUGGCUGAGCUUUGGUGGCGUCUGGCAGGCCGCUUCUGGCAGGCUGCGUCUGGCAGCAGUG-UUGAAGAUAUUUGUAACGAA----------- ...(..((...(((..((((((.(.....).))))))...)))..(((((.....)))))...)-)..)................----------- ( -24.60, z-score = 1.34, R) >droMoj3.scaffold_6473 8214113 85 + 16943266 UGAUUGGCUACGAGACGGCGGCGUCUGCCAGGCGGCUUCUGGCAGGCCGCUUCUGGCAACAGUACUUACAGUUUUAUGUAACGAG----------- ...(((..((((((((((((((..((((((((.....)))))))))))))).(((....)))........)))))..))).))).----------- ( -31.90, z-score = -1.70, R) >droGri2.scaffold_14853 1705925 84 + 10151454 UGAUUGGUUAAGACCAGGCCGCGUCUGGCAGGCUGCUUCUGGCAAGCCGCGUCUGGCAGUAGUG-CAGCAGGUAUUCGUAACGAU----------- ......((((..(((..((.(((.(((.(((((.((..((....))..))))))).)))...))-).)).))).....))))...----------- ( -25.40, z-score = 1.20, R) >droWil1.scaffold_181096 8544850 69 + 12416693 UGAUUGGUCGAGCUGUGGACGCGUCUGGCAGGCGGCAUCUGGCAGGUCGCGUCUGGCACUAGUGAUGAU--------------------------- ..((((((...(((..(((((((((((.((((.....)))).)))).))))))))))))))))......--------------------------- ( -27.50, z-score = -1.08, R) >consensus UGAUUGGCCAGAACAGGGCCGCGUCUGGCAGGCCACGUCUGGCAGGCCGCGUCUGGCAGCAGUGAUGACUGUUAU_GGCGACUCG___________ ..((((((((.......(((((..(((.((((.....)))).))))))))...))))..))))................................. (-16.61 = -15.40 + -1.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:18 2011