
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,522,801 – 4,522,886 |
| Length | 85 |
| Max. P | 0.957905 |

| Location | 4,522,801 – 4,522,886 |
|---|---|
| Length | 85 |
| Sequences | 4 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.46814 |
| G+C content | 0.48599 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951350 |
| Prediction | RNA |
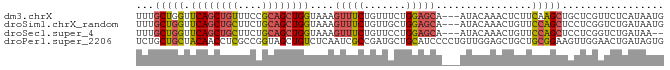
Download alignment: ClustalW | MAF
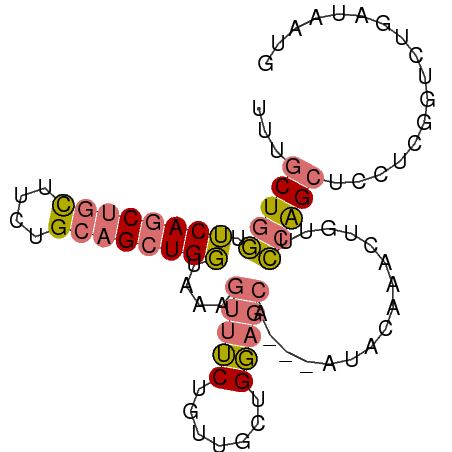
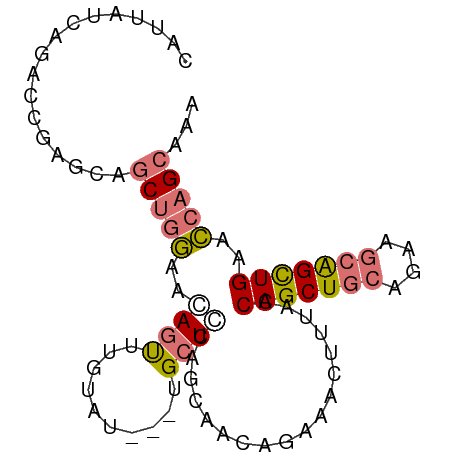
>dm3.chrX 4522801 85 + 22422827 UUUGCUGGUUCAGCUGUUUCCGCAGCUGGUAAAGUUUCUGUUUCUGGAGCA---AUACAAACUCUUCAAGCUGCUCGUUCUCAUAAUG ...(((...((((((((....))))))))...)))...........(((((---.................)))))............ ( -18.23, z-score = -0.37, R) >droSim1.chrX_random 1608518 85 + 5698898 UUUGCUGGUUCAGCUGCUUCUGCAGCUGGUAAAGUUUCUGUUGCUGGAGCA---AUACAAACUGUUCCAGCUCCUCGGUCUGAUAAUG ...(((...((((((((....))))))))...)))..(((..(((((((((---........)))))))))....))).......... ( -31.90, z-score = -3.70, R) >droSec1.super_4 3972919 83 - 6179234 UUUGCUGGUUCAGCUGCUUCUGCAGCUGGUAAAGUUUCUGUUCCUGGAGCA---AUACAAACUGUUCCAGCUCCUCGGUCUGAUAA-- ...(((...((((((((....))))))))...)))..(((...((((((((---........)))))))).....)))........-- ( -27.50, z-score = -2.62, R) >droPer1.super_2206 51 88 + 3766 UCUGCUGCUACAACCUCGCCGGUAGCUGUCUCAAUCGCCGAUGCUGCAUCCCCUGUUGGAGCUGCUGCGGAAGUUGGAACUGAUAGUG ...((((((...........)))))).((..((((..(((..((.((.(((......))))).))..)))..))))..))........ ( -24.70, z-score = 0.13, R) >consensus UUUGCUGGUUCAGCUGCUUCUGCAGCUGGUAAAGUUUCUGUUGCUGGAGCA___AUACAAACUGUUCCAGCUCCUCGGUCUGAUAAUG ...(((((.((((((((....))))))))....(((((.......)))))................)))))................. (-16.80 = -17.30 + 0.50)
| Location | 4,522,801 – 4,522,886 |
|---|---|
| Length | 85 |
| Sequences | 4 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.46814 |
| G+C content | 0.48599 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -13.66 |
| Energy contribution | -13.91 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957905 |
| Prediction | RNA |
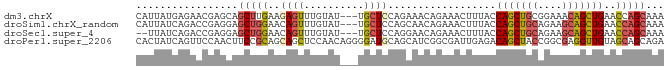
Download alignment: ClustalW | MAF
>dm3.chrX 4522801 85 - 22422827 CAUUAUGAGAACGAGCAGCUUGAAGAGUUUGUAU---UGCUCCAGAAACAGAAACUUUACCAGCUGCGGAAACAGCUGAACCAGCAAA ..............(((((((((((..(((((.(---(......)).)))))..)))))..))))))(....).((((...))))... ( -22.40, z-score = -2.35, R) >droSim1.chrX_random 1608518 85 - 5698898 CAUUAUCAGACCGAGGAGCUGGAACAGUUUGUAU---UGCUCCAGCAACAGAAACUUUACCAGCUGCAGAAGCAGCUGAACCAGCAAA .................(((((...(((((.(.(---(((....)))).).)))))....(((((((....)))))))..)))))... ( -29.50, z-score = -3.88, R) >droSec1.super_4 3972919 83 + 6179234 --UUAUCAGACCGAGGAGCUGGAACAGUUUGUAU---UGCUCCAGGAACAGAAACUUUACCAGCUGCAGAAGCAGCUGAACCAGCAAA --...............(((((...(((((.(.(---(.(....).)).).)))))....(((((((....)))))))..)))))... ( -25.00, z-score = -2.36, R) >droPer1.super_2206 51 88 - 3766 CACUAUCAGUUCCAACUUCCGCAGCAGCUCCAACAGGGGAUGCAGCAUCGGCGAUUGAGACAGCUACCGGCGAGGUUGUAGCAGCAGA ........(((.(((((((.((.(((.((((....)))).)))......(((..........)))....))))))))).)))...... ( -24.10, z-score = 0.24, R) >consensus CAUUAUCAGACCGAGCAGCUGGAACAGUUUGUAU___UGCUCCAGCAACAGAAACUUUACCAGCUGCAGAAGCAGCUGAACCAGCAAA .................(((((.....(((((...............)))))........(((((((....)))))))..)))))... (-13.66 = -13.91 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:15 2011