
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,481,881 – 4,481,971 |
| Length | 90 |
| Max. P | 0.786420 |

| Location | 4,481,881 – 4,481,971 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 62.95 |
| Shannon entropy | 0.69147 |
| G+C content | 0.52506 |
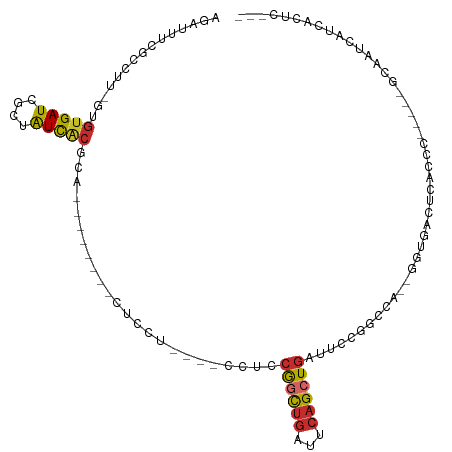
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -7.82 |
| Energy contribution | -8.04 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
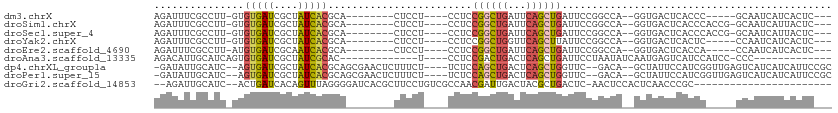
>dm3.chrX 4481881 90 + 22422827 AGAUUUCGCCUU-GUGUGAUCGCUAUCACGCA--------CUCCU----CCUCCGGCUGAUUCAGCUGAUUCCGGCCA--GGUGACUCACCC-----GCAAUCAUCACUC--- .((((.((((.(-(((((((....))))))))--------.....----....((((((...)))))).....)))..--((((...)))).-----).)))).......--- ( -25.40, z-score = -1.73, R) >droSim1.chrX 3484346 94 + 17042790 AGAUUUCGCCUU-GUGUGAUCGCUAUCACGCA--------CUCCU----CCUCCGGCUGAUUCAGCUGAUUCCGGCCA--GGUGACUCACCCACCG-GCAAUCAUUACUC--- .((((..(((.(-(((((((....))))))))--------.....----.....(((((..((....))...))))).--((((.......)))))-)))))).......--- ( -28.20, z-score = -2.35, R) >droSec1.super_4 3932802 94 - 6179234 AGAUUUCGCCUU-GUGUGAUCGCUAUCACGCA--------CUCCU----CCUCCGGCUGAUUCAGCUGAUUCCGGCCA--GGUGACUCACCCACCG-GCAAUCAUUACUC--- .((((..(((.(-(((((((....))))))))--------.....----.....(((((..((....))...))))).--((((.......)))))-)))))).......--- ( -28.20, z-score = -2.35, R) >droYak2.chrX 17814826 90 - 21770863 AGAUUUCGCCUU-GUGUGAUCGCUAUCACGCA--------CUCCU----CCUCCGGCUGGUUCAGCUUAUUCCGGCCA--GGUGACUCACUC-----CCAAUCAUCACUC--- .((..(((((((-(((((((....))))))))--------.....----.....((((((...........)))))))--))))).))....-----.............--- ( -26.40, z-score = -2.87, R) >droEre2.scaffold_4690 1861052 90 + 18748788 AGAUUUCGCCUU-AUGUGAUCGCAAUCACGCA--------CUCCU----CCUCCGGCUGAUUCAGCUGAUUCCGGCCA--GGUGACUCACCA-----CCAAUCAUCACUC--- .((((..(((..-..(((((....)))))...--------.....----....((((((...)))))).....)))..--((((......))-----)))))).......--- ( -22.80, z-score = -1.79, R) >droAna3.scaffold_13335 1445250 82 - 3335858 AGACAUUGCAUCAGUGUGAUCGCUAUCGCAC-------------U----CCUCCGACUGACUCAGCUGAUUCCUAAUAUCAAUGAGUCAUCCAUCC-CCC------------- ............((((((((....)))))))-------------)----.....((.(((((((..((((.......)))).))))))))).....-...------------- ( -20.60, z-score = -2.78, R) >dp4.chrXL_group1a 372992 102 - 9151740 -GAUAUUGCAUC--AGUGAUCGCUAUCACGCAGCGAACUCUUUCU----UCUCCAGCUGACUCAGCUGGUUC--GACA--GCUAUUCCAUCGGUUGAGUCAUCAUCAUUCCGC -(((.((((...--.(((((....)))))...)))).........----......(.(((((((((((((..--((..--.....)).))))))))))))).))))....... ( -26.00, z-score = -1.09, R) >droPer1.super_15 1009061 102 - 2181545 -GAUAUUGCAUC--AGUGAUCGCUAUCACGCAGCGAACUCUUUCU----UCUCCAGCUGACUCAGCUGGUUC--GACA--GCUAUUCCAUCGGUUGAGUCAUCAUCAUUCCGC -(((.((((...--.(((((....)))))...)))).........----......(.(((((((((((((..--((..--.....)).))))))))))))).))))....... ( -26.00, z-score = -1.09, R) >droGri2.scaffold_14853 1234269 85 + 10151454 --AGAUUGCAUC--ACUGAUCACAGUUUAGGGGAUCACGCUUCCUGUCGCCAACGAUUGACUACGCUGACUC-AACUCCACUCAACCCGC----------------------- --.((((.....--...)))).((((.(((((((.......))))((((....))))...))).))))....-.................----------------------- ( -14.10, z-score = 0.70, R) >consensus AGAUUUCGCCUU_GUGUGAUCGCUAUCACGCA________CUCCU____CCUCCGGCUGAUUCAGCUGAUUCCGGCCA__GGUGACUCACCC_____GCAAUCAUCACUC___ ...............(((((....)))))........................((((((...))))))............................................. ( -7.82 = -8.04 + 0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:11 2011