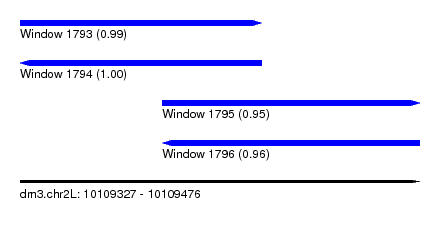
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,109,327 – 10,109,476 |
| Length | 149 |
| Max. P | 0.998390 |

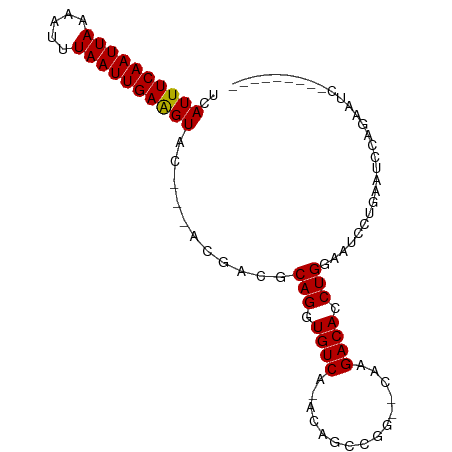
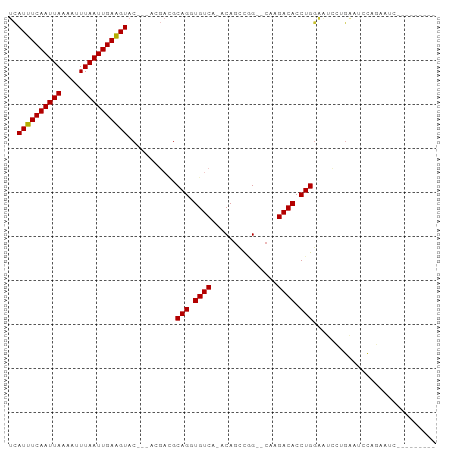
| Location | 10,109,327 – 10,109,417 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.77 |
| Shannon entropy | 0.49686 |
| G+C content | 0.42064 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.47 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
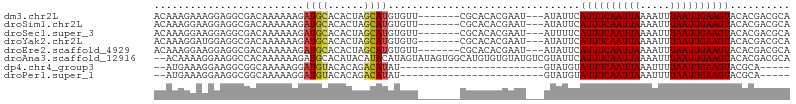
>dm3.chr2L 10109327 90 + 23011544 -AGAGACUGGAUUCAGGAUUCAGGAAUCCAGGUGUCUUG--CCGGCUGU-UGACACCUGCGUCGU---GUACUUCAAUUAAAUUUUAAUUGAAAUGA -...(((.((((((.........))))))(((((((..(--(.....))-.)))))))..)))..---....((((((((.....)))))))).... ( -25.40, z-score = -1.56, R) >droSim1.chr2L 9900501 82 + 22036055 ---------GAGACUGGAUUCAGGAUUCCAGGUGUCUUG--CCGGCUGU-UGACACCUGCGUCGU---GUACUUCAAUUAAAUUUUAAUUGAAAUGA ---------...........((.(((..((((((((..(--(.....))-.)))))))).))).)---)...((((((((.....)))))))).... ( -21.70, z-score = -1.40, R) >droSec1.super_3 5547139 82 + 7220098 ---------GAGACUGGAUUCAGGAUUCCAGGUGUCUUG--CCGGCUGU-UGACACCUGCGUCGU---GUACUUCAAUUAAAUUUUAAUUGAAAUGA ---------...........((.(((..((((((((..(--(.....))-.)))))))).))).)---)...((((((((.....)))))))).... ( -21.70, z-score = -1.40, R) >droYak2.chr2L 12793282 91 + 22324452 AGGAGGCAGGAUUCAGGAUUCAGAAUUCCAGGUGUCUUG--CCGACUGU-UGACACCUGCGUCGU---GUACUUCAAUUAAAUUUUAAUUGAAAUGA ((..((((((((.(.(((........)))..).))))))--))..))..-.(((......)))..---....((((((((.....)))))))).... ( -24.90, z-score = -1.34, R) >droEre2.scaffold_4929 10724585 91 + 26641161 GAGACCGGGGAUUCAGGAUUCAGGAUUCCAGGUGUCUUG--CCGGCUGU-UGACACCUGCGUCGU---GUACUUCAAUUAAAUUUUAAUUGAAAUGA ..((((.(((((((........)))))))(((((((..(--(.....))-.)))))))).)))..---....((((((((.....)))))))).... ( -25.90, z-score = -1.06, R) >droAna3.scaffold_12916 13942510 68 + 16180835 -----------------------GUUUCCAGGUGUCUUG--CCGGCUGU-UGACAUCUGCGUCGU---GUACUUCAAUUAAAUUUUAAUUGAAAUGA -----------------------.....((((((((..(--(.....))-.))))))))......---....((((((((.....)))))))).... ( -14.90, z-score = -1.44, R) >dp4.chr4_group3 11609438 75 - 11692001 ----------GCCAUGUACACCCGUGGCCAGGUGUCUUG--C-GGCUGU-UGACAUCUGC--------GUACUUCAAUUAAAAUUUAAUUGAAAUAC ----------((((((......))))))((((((((..(--(-....))-.)))))))).--------(((.((((((((.....)))))))).))) ( -24.90, z-score = -3.57, R) >droPer1.super_1 8726356 75 - 10282868 ----------GCCAUGUACACCCGUGGCCAGGUGUCUUG--C-GGCUGU-UGACAUCUGC--------GUACUUCAAUUAAAAUUUAAUUGAAAUAC ----------((((((......))))))((((((((..(--(-....))-.)))))))).--------(((.((((((((.....)))))))).))) ( -24.90, z-score = -3.57, R) >droWil1.scaffold_181132 86985 85 + 1035393 ----------GUCACACAUACUGAGA--CAGGUGUCUUUGCCUAGUUGUAUGACAUCUGCAUCGUCAUGUACCUCAAUUAAAUUUUAAUUGAAAUGA ----------.....((((..(((..--((((((((..(((......))).))))))))..)))..))))...(((((((.....)))))))..... ( -21.10, z-score = -1.92, R) >consensus _________GAUCCUGGAUUCAGGAUUCCAGGUGUCUUG__CCGGCUGU_UGACACCUGCGUCGU___GUACUUCAAUUAAAUUUUAAUUGAAAUGA ............................((((((((...............)))))))).............((((((((.....)))))))).... (-15.61 = -15.47 + -0.14)
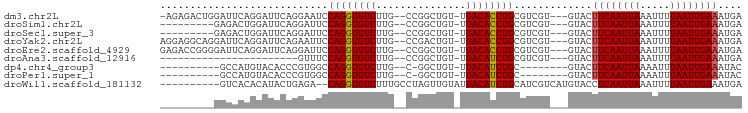
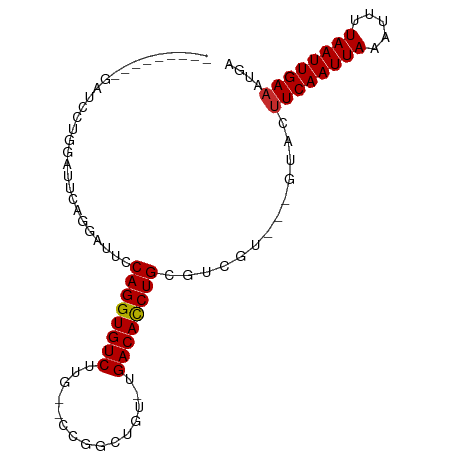
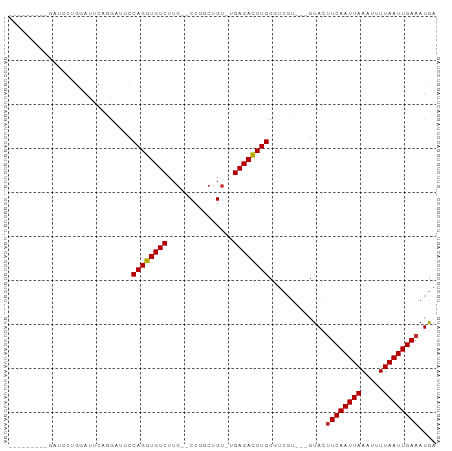
| Location | 10,109,327 – 10,109,417 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.77 |
| Shannon entropy | 0.49686 |
| G+C content | 0.42064 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -12.55 |
| Energy contribution | -12.45 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10109327 90 - 23011544 UCAUUUCAAUUAAAAUUUAAUUGAAGUAC---ACGACGCAGGUGUCA-ACAGCCGG--CAAGACACCUGGAUUCCUGAAUCCUGAAUCCAGUCUCU- ..((((((((((.....))))))))))..---..(((..(((((((.-........--...)))))))((((((..(....).)))))).)))...- ( -26.20, z-score = -3.55, R) >droSim1.chr2L 9900501 82 - 22036055 UCAUUUCAAUUAAAAUUUAAUUGAAGUAC---ACGACGCAGGUGUCA-ACAGCCGG--CAAGACACCUGGAAUCCUGAAUCCAGUCUC--------- ..((((((((((.....))))))))))..---..(((..(((((((.-........--...)))))))(((........))).)))..--------- ( -21.90, z-score = -2.99, R) >droSec1.super_3 5547139 82 - 7220098 UCAUUUCAAUUAAAAUUUAAUUGAAGUAC---ACGACGCAGGUGUCA-ACAGCCGG--CAAGACACCUGGAAUCCUGAAUCCAGUCUC--------- ..((((((((((.....))))))))))..---..(((..(((((((.-........--...)))))))(((........))).)))..--------- ( -21.90, z-score = -2.99, R) >droYak2.chr2L 12793282 91 - 22324452 UCAUUUCAAUUAAAAUUUAAUUGAAGUAC---ACGACGCAGGUGUCA-ACAGUCGG--CAAGACACCUGGAAUUCUGAAUCCUGAAUCCUGCCUCCU ..((((((((((.....))))))))))..---..((.(((((..(((-...(((..--...)))....(((........))))))..))))).)).. ( -23.40, z-score = -2.57, R) >droEre2.scaffold_4929 10724585 91 - 26641161 UCAUUUCAAUUAAAAUUUAAUUGAAGUAC---ACGACGCAGGUGUCA-ACAGCCGG--CAAGACACCUGGAAUCCUGAAUCCUGAAUCCCCGGUCUC ..((((((((((.....))))))))))..---..(((..(((((((.-........--...)))))))(((.((..(....).)).)))...))).. ( -21.40, z-score = -1.46, R) >droAna3.scaffold_12916 13942510 68 - 16180835 UCAUUUCAAUUAAAAUUUAAUUGAAGUAC---ACGACGCAGAUGUCA-ACAGCCGG--CAAGACACCUGGAAAC----------------------- ..((((((((((.....))))))))))..---......(((.((((.-........--...)))).))).....----------------------- ( -12.90, z-score = -1.94, R) >dp4.chr4_group3 11609438 75 + 11692001 GUAUUUCAAUUAAAUUUUAAUUGAAGUAC--------GCAGAUGUCA-ACAGCC-G--CAAGACACCUGGCCACGGGUGUACAUGGC---------- ((((((((((((.....))))))))))))--------..........-...(((-(--....(((((((....)))))))...))))---------- ( -26.90, z-score = -4.34, R) >droPer1.super_1 8726356 75 + 10282868 GUAUUUCAAUUAAAUUUUAAUUGAAGUAC--------GCAGAUGUCA-ACAGCC-G--CAAGACACCUGGCCACGGGUGUACAUGGC---------- ((((((((((((.....))))))))))))--------..........-...(((-(--....(((((((....)))))))...))))---------- ( -26.90, z-score = -4.34, R) >droWil1.scaffold_181132 86985 85 - 1035393 UCAUUUCAAUUAAAAUUUAAUUGAGGUACAUGACGAUGCAGAUGUCAUACAACUAGGCAAAGACACCUG--UCUCAGUAUGUGUGAC---------- ..((((((((((.....))))))))))................((((((((((((((((........))--))).))).))))))))---------- ( -21.80, z-score = -2.07, R) >consensus UCAUUUCAAUUAAAAUUUAAUUGAAGUAC___ACGACGCAGGUGUCA_ACAGCCGG__CAAGACACCUGGAAUCCUGAAUCCAGAAUC_________ ..((((((((((.....))))))))))...........(((.((((...............)))).)))............................ (-12.55 = -12.45 + -0.10)

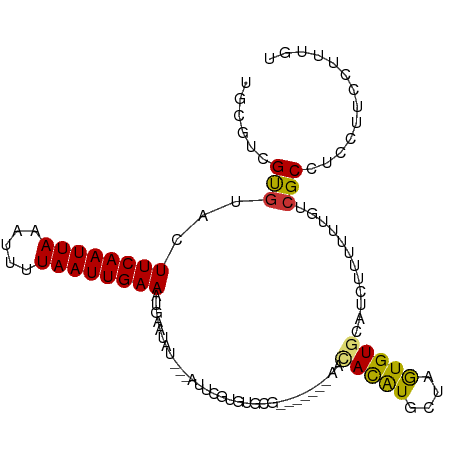
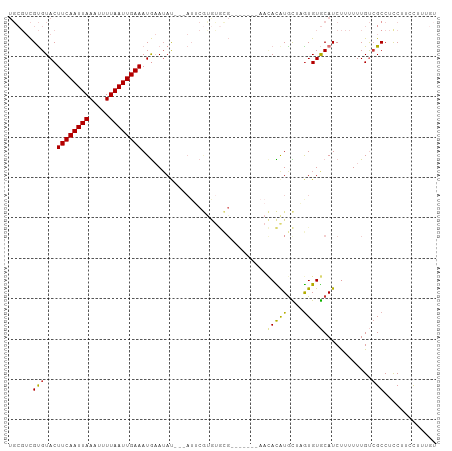
| Location | 10,109,380 – 10,109,476 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.12 |
| Shannon entropy | 0.45644 |
| G+C content | 0.34993 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -8.12 |
| Energy contribution | -7.78 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
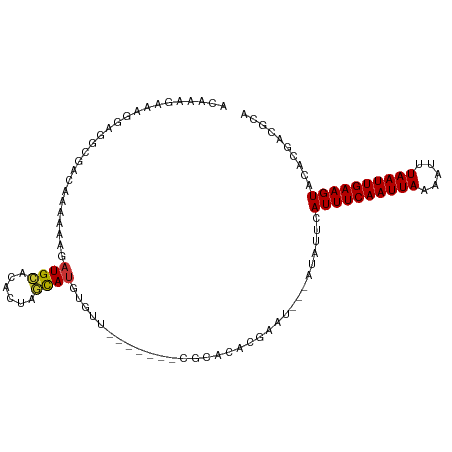
>dm3.chr2L 10109380 96 + 23011544 UGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUAU---AUUCGUGUGCG-------AACACAUGCUAGUGUGCAUCUUUUUUGUCGCCUCCUUUCUUUGU .((((((..(((((((((((.....))))))))..(((...---.))))))..))-------).(((((....)))))............)))............. ( -19.90, z-score = -1.91, R) >droSim1.chr2L 9900546 96 + 22036055 UGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUAU---AUUCGUGUGCG-------AACACAUGCUAGUGUGCAUCUUUUUUGUCGCCUCCUUCCUUUGU .((((((..(((((((((((.....))))))))..(((...---.))))))..))-------).(((((....)))))............)))............. ( -19.90, z-score = -2.08, R) >droSec1.super_3 5547184 96 + 7220098 UGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAAAU---AUUCGUGUGCG-------AACACAUGCUAGUGUGCAUCUUUUUUGUCGCCUCCUUCCUUUGU .((((((..(((((((((((.....))))))))..(((...---.))))))..))-------).(((((....)))))............)))............. ( -19.90, z-score = -2.11, R) >droYak2.chr2L 12793336 96 + 22324452 UGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUAU---AUUCGUGUGCG-------CACACAUGCUAGUGUGCAUCUUUUUUGUCGCCUCCAUCCUUUGU .(((.((.((((((((((((.....))))))))(((((...---.)))))))))(-------(((((......))))))........)).)))............. ( -24.50, z-score = -3.23, R) >droEre2.scaffold_4929 10724639 96 + 26641161 UGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUAU---AUUCGUGUGCG-------AACACAUGCUAGUGUGCAUCUUUUUUGUCGCCUCCUUCCUUUGU .((((((..(((((((((((.....))))))))..(((...---.))))))..))-------).(((((....)))))............)))............. ( -19.90, z-score = -2.08, R) >droAna3.scaffold_12916 13942541 104 + 16180835 UGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUACGACAUACACACAUGCCACUAUACUAUGUAUGUAUGUGCAUCUUUUUUGUGGCCUUCCUUUUGU-- ...(((((((.(((((((((.....))))))))..).)))))))..........(((((......((((((....)))))).......)))))...........-- ( -25.22, z-score = -3.00, R) >dp4.chr4_group3 11609481 75 - 11692001 -----UGCGUACUUCAAUUAAAAUUUAAUUGAAAUACAUAC------------------------AUAUGUCUGUGUACAUCCUUUUUGCCGCCUUCCUUUCAU-- -----.(((((.((((((((.....)))))))).))).(((------------------------(((....)))))).............))...........-- ( -12.70, z-score = -3.45, R) >droPer1.super_1 8726399 75 - 10282868 -----UGCGUACUUCAAUUAAAAUUUAAUUGAAAUACAUAC------------------------AUAUGUCUGUGUACAUCCUUUUUGCCGCCUUCCUUUCAU-- -----.(((((.((((((((.....)))))))).))).(((------------------------(((....)))))).............))...........-- ( -12.70, z-score = -3.45, R) >consensus UGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUAU___AUUCGUGUGCG_______AACACAUGCUAGUGUGCAUCUUUUUUGUCGCCUCCUUCCUUUGU ......(((...((((((((.....))))))))...............................(((((....)))))............)))............. ( -8.12 = -7.78 + -0.34)

| Location | 10,109,380 – 10,109,476 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.12 |
| Shannon entropy | 0.45644 |
| G+C content | 0.34993 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -8.70 |
| Energy contribution | -8.53 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10109380 96 - 23011544 ACAAAGAAAGGAGGCGACAAAAAAGAUGCACACUAGCAUGUGUU-------CGCACACGAAU---AUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCA .............(((........(((((......)))((((((-------((....)))))---))).))((((((((((.....))))))))))......))). ( -19.70, z-score = -2.49, R) >droSim1.chr2L 9900546 96 - 22036055 ACAAAGGAAGGAGGCGACAAAAAAGAUGCACACUAGCAUGUGUU-------CGCACACGAAU---AUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCA .............(((........(((((......)))((((((-------((....)))))---))).))((((((((((.....))))))))))......))). ( -19.70, z-score = -2.37, R) >droSec1.super_3 5547184 96 - 7220098 ACAAAGGAAGGAGGCGACAAAAAAGAUGCACACUAGCAUGUGUU-------CGCACACGAAU---AUUUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCA .............(((.........((((......))))(((((-------((....)))))---))....((((((((((.....))))))))))......))). ( -19.00, z-score = -1.92, R) >droYak2.chr2L 12793336 96 - 22324452 ACAAAGGAUGGAGGCGACAAAAAAGAUGCACACUAGCAUGUGUG-------CGCACACGAAU---AUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCA .............(((........(.(((((((......)))))-------)))........---......((((((((((.....))))))))))......))). ( -21.90, z-score = -2.44, R) >droEre2.scaffold_4929 10724639 96 - 26641161 ACAAAGGAAGGAGGCGACAAAAAAGAUGCACACUAGCAUGUGUU-------CGCACACGAAU---AUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCA .............(((........(((((......)))((((((-------((....)))))---))).))((((((((((.....))))))))))......))). ( -19.70, z-score = -2.37, R) >droAna3.scaffold_12916 13942541 104 - 16180835 --ACAAAAGGAAGGCCACAAAAAAGAUGCACAUACAUACAUAGUAUAGUGGCAUGUGUGUAUGUCGUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCA --......((....))..........(((..((((((((((.((......))))))))))))(((((....((((((((((.....))))))))))..)))))))) ( -27.40, z-score = -3.46, R) >dp4.chr4_group3 11609481 75 + 11692001 --AUGAAAGGAAGGCGGCAAAAAGGAUGUACACAGACAUAU------------------------GUAUGUAUUUCAAUUAAAUUUUAAUUGAAGUACGCA----- --..............((.........(((((........)------------------------))))((((((((((((.....)))))))))))))).----- ( -15.90, z-score = -2.94, R) >droPer1.super_1 8726399 75 + 10282868 --AUGAAAGGAAGGCGGCAAAAAGGAUGUACACAGACAUAU------------------------GUAUGUAUUUCAAUUAAAUUUUAAUUGAAGUACGCA----- --..............((.........(((((........)------------------------))))((((((((((((.....)))))))))))))).----- ( -15.90, z-score = -2.94, R) >consensus ACAAAGAAAGGAGGCGACAAAAAAGAUGCACACUAGCAUGUGUU_______CGCACACGAAU___AUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCA .........................((((......))))................................((((((((((.....)))))))))).......... ( -8.70 = -8.53 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:37 2011