
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,459,620 – 4,459,687 |
| Length | 67 |
| Max. P | 0.577218 |

| Location | 4,459,620 – 4,459,687 |
|---|---|
| Length | 67 |
| Sequences | 8 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 68.76 |
| Shannon entropy | 0.57087 |
| G+C content | 0.52301 |
| Mean single sequence MFE | -21.81 |
| Consensus MFE | -9.99 |
| Energy contribution | -9.90 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4459620 67 + 22422827 UUUUUGGAUACCUAUGCGGGUCCCAUGGAAAGCUCGGGUGAUAAGCGUU--------------UGCU-GCGCGUGGGAAAAU ..........((((((((.(((((.(((.....))))).))).(((...--------------.)))-.))))))))..... ( -18.30, z-score = -0.12, R) >droSim1.chrX_random 1600562 67 + 5698898 UUUUUGGAUACCUAUGCGGGUCCCAUGGAAAGCUUAGGUGAUAAGCGUU--------------UGCU-GGGCGUGGGAAAAU ..........(((((((.....(((.(.((((((((.....))))).))--------------).))-)))))))))..... ( -18.20, z-score = -0.71, R) >droSec1.super_4 3910708 68 - 6179234 UUUUUGGAUACCUAUGCGGGUCCCAUGGAAAGCUUUGGUGAUAAGCGUU--------------UGCUUGGGCGUGGGAAAAU ..........(((((((..((((((.((....)).))).)))((((...--------------.))))..)))))))..... ( -19.00, z-score = -0.91, R) >droYak2.chrX 17793257 67 - 21770863 UUUUUGGAGACCCAUGCGGGUCCCAUGAAAAGCUUUGGUGAUAAGCGUU---------------UUCUGGGCGUGGGAAAAU ..........(((((((.....(((.(((((((((.......)))).))---------------)))))))))))))..... ( -20.90, z-score = -1.35, R) >droEre2.scaffold_4690 1838874 71 + 18748788 UUUUUGGAGACCCAUGCGGGUCCCAUGAAAAGCUUCGAUGAUAAGCGUUG-----------CGUUGCUGGGCGUGGGAAAAU (((.(((.(((((....)))))))).)))....(((.(((...((((...-----------...))))...))).))).... ( -21.90, z-score = -0.94, R) >dp4.chrXL_group1a 345120 79 - 9151740 -UUUUGGGAUCCCAUGCGGG--CGGUGAAAAGCUCCAGCGAUAAGCGGCGGCUUGGUGGGGCAGCUGGGGGCGUGGGAAAAC -........((((((((...--((((.....((((((....(((((....))))).)))))).))))...)))))))).... ( -30.50, z-score = -1.60, R) >droPer1.super_15 981143 79 - 2181545 -UUUUGGGAUCCCAUGCGGG--CGGUGAAAAGCUCCAGCGAUAAACGGCGGCUUGGUGGGGCAGCUGGGGGCGUGGGAAAAC -........(((((((((((--(........)).)).........((((.(((......))).))))...)))))))).... ( -28.80, z-score = -1.36, R) >droVir3.scaffold_13042 2893609 53 + 5191987 ---------GUCUAUGCGGG---CGCGAAAAGCUCCAAAGAUAAGUUUU-----------------UGGGGCGUGGGAAAAU ---------.(((((((...---.)).....((((((((((....))))-----------------)))))))))))..... ( -16.90, z-score = -2.44, R) >consensus UUUUUGGAUACCCAUGCGGGUCCCAUGAAAAGCUCCGGUGAUAAGCGUU______________UGCUGGGGCGUGGGAAAAU ..........((((((((.....).......(((.........)))........................)))))))..... ( -9.99 = -9.90 + -0.09)

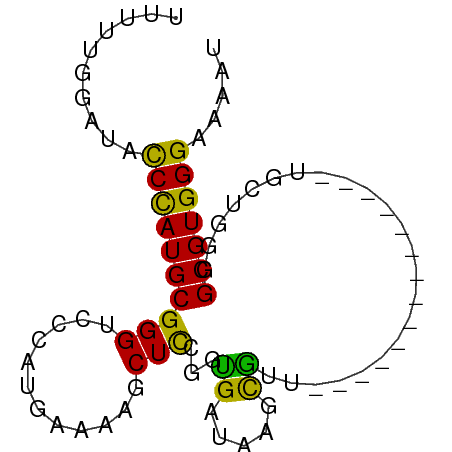
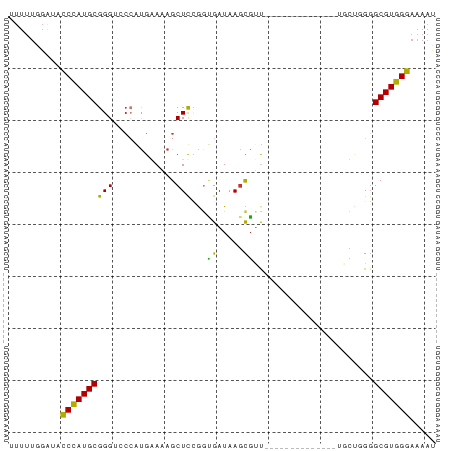
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:06 2011