
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,457,307 – 4,457,403 |
| Length | 96 |
| Max. P | 0.887892 |

| Location | 4,457,307 – 4,457,403 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.17 |
| Shannon entropy | 0.48284 |
| G+C content | 0.56671 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.65 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
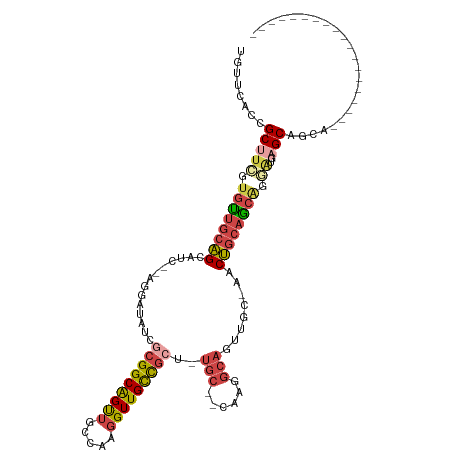
>dm3.chrX 4457307 96 + 22422827 UGUUCACCGCAUCGUGUUGCAGCAUC--AGGAUAUCGCGGCAGUUGCCAAGGUUGCCGCUCUGC--CAAGGCAGUUGC-AACUGCAGCGGGAUAAGCAGCA------------------- ((((..((((..((.((((((((...--........((((((...((....))))))))..(((--....))))))))-)))))..))))....))))...------------------- ( -35.90, z-score = -0.53, R) >droSim1.chrX 3458661 95 + 17042790 UGUUCACCGCUUCGUGUUGCAGCAUC--AGGAUAUCGCGGCAGUUGCCAAGGUUGCCGCU-UGC--CAAGGCAGUUGC-AACUGCAGCGGGAUGAGCAGCA------------------- (((((((((((.((.((((((((...--........((((((...((....)))))))).-(((--....))))))))-))))).)))))..))))))...------------------- ( -42.60, z-score = -2.25, R) >droSec1.super_4 3908363 95 - 6179234 UGUUCACCGCUUCGUGUUGCAGCAUC--AGGAUAUCGCGGCAGUUUCCAAGGUUGCCGCU-UGC--AAAGGCAGUUGC-AACUGCAGCGGGAUGAGCAGCA------------------- (((((((((((.((.((((((((...--........(((((((..(...)..))))))).-(((--....))))))))-))))).)))))..))))))...------------------- ( -41.00, z-score = -2.63, R) >droYak2.chrX 17790429 95 - 21770863 UGUUGACCGCUAUGUGCUGCAGCAUC--AGGAUAUCGCGGCAGUUGCCAAGGUUGCCGAG-UGC--CCGAGCAGUUGC-AACUGCAGCAGAAUGAGCAGCA------------------- .((((.((((.((((.(((......)--)).)))).))))..(((((...((((((.((.-(((--....))).))))-)))))))))........)))).------------------- ( -34.00, z-score = -0.05, R) >droEre2.scaffold_4690 1836517 95 + 18748788 UGUUGGCCGCUUGGUGUUGCAGCAUC--AGGAUAUCGCGGCAGUUGCCAAGGUUGCCGCU-UGC--CAAGGCAGUUGC-AACUGCAGCAGGAUGAGCAGCA------------------- .((((((((((((((((....)))))--))(....)))))).(((((...((((((.(((-.((--....))))).))-)))))))))........)))).------------------- ( -38.70, z-score = -0.33, R) >droAna3.scaffold_13335 1416867 116 - 3335858 ----UGCAGCAUCGCUUCCCUGCCUCCAAGGACGGACGGGCGGCCGUUGGUGUUGUUGUUCUUCUUCCAGAUCCUGGCUAACAACAACACCGUUGGCCGCCCUAAUACACUUCCUUAUAA ----.((((..........))))....(((((.(...(((((((((.(((((((((((((......((((...))))..))))))))))))).))))))))).......).))))).... ( -48.70, z-score = -5.29, R) >consensus UGUUCACCGCUUCGUGUUGCAGCAUC__AGGAUAUCGCGGCAGUUGCCAAGGUUGCCGCU_UGC__CAAGGCAGUUGC_AACUGCAGCAGGAUGAGCAGCA___________________ ........(((((.((((((((..............(((((((((.....)))))))))...........((....))...)))))))).))..)))....................... (-19.84 = -20.65 + 0.81)

| Location | 4,457,307 – 4,457,403 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.17 |
| Shannon entropy | 0.48284 |
| G+C content | 0.56671 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -20.76 |
| Energy contribution | -19.77 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
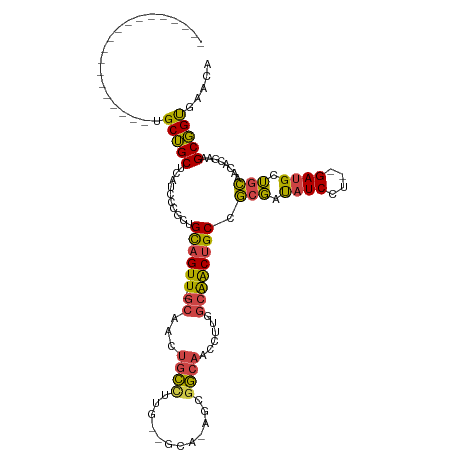
>dm3.chrX 4457307 96 - 22422827 -------------------UGCUGCUUAUCCCGCUGCAGUU-GCAACUGCCUUG--GCAGAGCGGCAACCUUGGCAACUGCCGCGAUAUCCU--GAUGCUGCAACACGAUGCGGUGAACA -------------------............((((((((((-(((.((((....--)))).((((((.....(....)))))))........--.....))))))....))))))).... ( -34.50, z-score = -1.25, R) >droSim1.chrX 3458661 95 - 17042790 -------------------UGCUGCUCAUCCCGCUGCAGUU-GCAACUGCCUUG--GCA-AGCGGCAACCUUGGCAACUGCCGCGAUAUCCU--GAUGCUGCAACACGAAGCGGUGAACA -------------------(((.(((((...(((.((((((-((....((....--))(-((.(....)))).)))))))).)))......)--)).)).))).(((......))).... ( -32.50, z-score = -0.56, R) >droSec1.super_4 3908363 95 + 6179234 -------------------UGCUGCUCAUCCCGCUGCAGUU-GCAACUGCCUUU--GCA-AGCGGCAACCUUGGAAACUGCCGCGAUAUCCU--GAUGCUGCAACACGAAGCGGUGAACA -------------------.........(((((((...(((-(((..(((....--)))-.((((((.....(....)))))))........--.....))))))....))))).))... ( -30.80, z-score = -1.04, R) >droYak2.chrX 17790429 95 + 21770863 -------------------UGCUGCUCAUUCUGCUGCAGUU-GCAACUGCUCGG--GCA-CUCGGCAACCUUGGCAACUGCCGCGAUAUCCU--GAUGCUGCAGCACAUAGCGGUCAACA -------------------.((((((.....(((((((((.-((....))((((--(..-(.(((((.....(....)))))).)....)))--)).)))))))))...))))))..... ( -34.80, z-score = -1.21, R) >droEre2.scaffold_4690 1836517 95 - 18748788 -------------------UGCUGCUCAUCCUGCUGCAGUU-GCAACUGCCUUG--GCA-AGCGGCAACCUUGGCAACUGCCGCGAUAUCCU--GAUGCUGCAACACCAAGCGGCCAACA -------------------.((((((.....((.((((((.-.((..(((....--)))-.((((((.....(....))))))).......)--)..)))))).))...))))))..... ( -32.40, z-score = -0.48, R) >droAna3.scaffold_13335 1416867 116 + 3335858 UUAUAAGGAAGUGUAUUAGGGCGGCCAACGGUGUUGUUGUUAGCCAGGAUCUGGAAGAAGAACAACAACACCAACGGCCGCCCGUCCGUCCUUGGAGGCAGGGAAGCGAUGCUGCA---- ..........(((((((.((((((((...((((((((((((..((((...))))......))))))))))))...))))))))((...((((((....)))))).))))))).)).---- ( -54.00, z-score = -5.09, R) >consensus ___________________UGCUGCUCAUCCCGCUGCAGUU_GCAACUGCCUUG__GCA_AGCGGCAACCUUGGCAACUGCCGCGAUAUCCU__GAUGCUGCAACACGAAGCGGUGAACA ....................(((((.....((((((((((.....)))))..........))))).......(((....)))(((.((((....)))).)))........)))))..... (-20.76 = -19.77 + -0.99)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:06 2011