
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,428,229 – 4,428,339 |
| Length | 110 |
| Max. P | 0.928176 |

| Location | 4,428,229 – 4,428,339 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Shannon entropy | 0.38655 |
| G+C content | 0.41079 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -16.22 |
| Energy contribution | -17.25 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
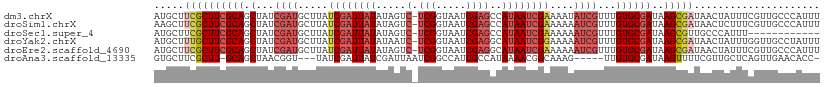
>dm3.chrX 4428229 110 + 22422827 AUGCUUCGCUUCGCAGCUAUCGAUGCUUAUCGAUUAUAUAGUC-UCGGUAAUCGAGCCAUAAUCGAAAAUAUCGUUUGUGCGAUAAGCGAUAACUAUUUCGUUGCCCAUUU .....((((((((((.(...(((((.((.((((((((...(.(-(((.....))))).)))))))).)))))))...))))))..)))))((((......))))....... ( -29.30, z-score = -1.65, R) >droSim1.chrX 3427812 110 + 17042790 AAGCUUCGCUUCGCAGCUAUCGAUGCUUAUCGAUUAUAUAGUC-UCGGUAAUCGAGCCAUAAUCGAAAAAAUCGUUUGUGCGAUAAGCGAUAACUCUUUCGUUGCCCAUUU ..((.((((((((((.(...((((..((.((((((((...(.(-(((.....))))).)))))))).)).))))...))))))..))))).(((......)))))...... ( -28.70, z-score = -1.64, R) >droSec1.super_4 3879932 98 - 6179234 AUGCUUCGCUUCGCAGCUAUCGAUGCUUAUCGAUUAUAUAGUC-UCGGUAAUCGAGCCAUAAUCGAAAAAAUCGUUUGUGCGAUAAGCGUUGCCCAUUU------------ ..((..(((((((((.(...((((..((.((((((((...(.(-(((.....))))).)))))))).)).))))...))))))..))))..))......------------ ( -29.10, z-score = -2.37, R) >droYak2.chrX 17761408 110 - 21770863 AUGCUUUGCUUCGCAGCUAUCGAUGCUUAUCGAUUAUAUAAUC-UCGGUAAUCGAGGCAUAAUCGGAAAAAUCGUUUGUGCGAUAAGCGAUAACUAUUUGGUUGCCUAUUU ............(((((((..((((.((((((.((((.....(-(((.....))))((((((.(((.....))).)))))).)))).)))))).)))))))))))...... ( -28.50, z-score = -1.16, R) >droEre2.scaffold_4690 1806907 110 + 18748788 AUGCUUCGCUUCGCAGCUAUCGAUGCUUAUCGAUUAUAUAGUC-UCGGUAAUCGAGGCAUAAUCGAAAAAAUCGUUUGUGCGAUAAGCGAUAACUAUUUCGUUGCCCAUUU .....((((((((((.(...((((..((.((((((((...(((-(((.....)))))))))))))).)).))))...))))))..)))))((((......))))....... ( -32.70, z-score = -2.45, R) >droAna3.scaffold_13335 3323354 101 - 3335858 GUGCUUCGCUU-GCAGCUAACGGU---UAUCGAUUAUCGAUUAAUCGGCCAUCGCCAUAAAACGGCAAAG-----UUGUGCGAUAAGUUUUCGUUGCUCAGUUGAACACC- (((.((((((.-(((((.((((((---...((((((.....)))))))))(((((.....(((......)-----))..)))))..)))...)))))..)).))))))).- ( -26.90, z-score = -0.99, R) >consensus AUGCUUCGCUUCGCAGCUAUCGAUGCUUAUCGAUUAUAUAGUC_UCGGUAAUCGAGCCAUAAUCGAAAAAAUCGUUUGUGCGAUAAGCGAUAACUAUUUCGUUGCCCAUUU .....((((((((((....(((((.....((((((((..........))))))))......)))))(((.....))).)))))..)))))..................... (-16.22 = -17.25 + 1.03)

| Location | 4,428,229 – 4,428,339 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Shannon entropy | 0.38655 |
| G+C content | 0.41079 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -14.81 |
| Energy contribution | -16.67 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4428229 110 - 22422827 AAAUGGGCAACGAAAUAGUUAUCGCUUAUCGCACAAACGAUAUUUUCGAUUAUGGCUCGAUUACCGA-GACUAUAUAAUCGAUAAGCAUCGAUAGCUGCGAAGCGAAGCAU ......(((((......))).(((((..((((((...((((.((.(((((((((.((((.....)))-)....))))))))).))..))))...).)))))))))).)).. ( -33.40, z-score = -3.20, R) >droSim1.chrX 3427812 110 - 17042790 AAAUGGGCAACGAAAGAGUUAUCGCUUAUCGCACAAACGAUUUUUUCGAUUAUGGCUCGAUUACCGA-GACUAUAUAAUCGAUAAGCAUCGAUAGCUGCGAAGCGAAGCUU ....((((..(....).....(((((..((((((...(((((((.(((((((((.((((.....)))-)....))))))))).))).))))...).)))))))))).)))) ( -35.00, z-score = -3.40, R) >droSec1.super_4 3879932 98 + 6179234 ------------AAAUGGGCAACGCUUAUCGCACAAACGAUUUUUUCGAUUAUGGCUCGAUUACCGA-GACUAUAUAAUCGAUAAGCAUCGAUAGCUGCGAAGCGAAGCAU ------------......((..((((..((((((...(((((((.(((((((((.((((.....)))-)....))))))))).))).))))...).)))))))))..)).. ( -32.40, z-score = -3.89, R) >droYak2.chrX 17761408 110 + 21770863 AAAUAGGCAACCAAAUAGUUAUCGCUUAUCGCACAAACGAUUUUUCCGAUUAUGCCUCGAUUACCGA-GAUUAUAUAAUCGAUAAGCAUCGAUAGCUGCGAAGCAAAGCAU .....(....).....((((((((((((((........((....)).(((((((.((((.....)))-)....))))))))))))))...)))))))((........)).. ( -25.40, z-score = -2.01, R) >droEre2.scaffold_4690 1806907 110 - 18748788 AAAUGGGCAACGAAAUAGUUAUCGCUUAUCGCACAAACGAUUUUUUCGAUUAUGCCUCGAUUACCGA-GACUAUAUAAUCGAUAAGCAUCGAUAGCUGCGAAGCGAAGCAU ......(((((......))).(((((..((((((...(((((((.(((((((((.((((.....)))-)....))))))))).))).))))...).)))))))))).)).. ( -31.40, z-score = -2.72, R) >droAna3.scaffold_13335 3323354 101 + 3335858 -GGUGUUCAACUGAGCAACGAAAACUUAUCGCACAA-----CUUUGCCGUUUUAUGGCGAUGGCCGAUUAAUCGAUAAUCGAUA---ACCGUUAGCUGC-AAGCGAAGCAC -(.(((((....))))).).........((((....-----..(((((((...))))))).(((..((..((((.....)))).---...))..)))..-..))))..... ( -23.00, z-score = -0.06, R) >consensus AAAUGGGCAACGAAAUAGUUAUCGCUUAUCGCACAAACGAUUUUUUCGAUUAUGGCUCGAUUACCGA_GACUAUAUAAUCGAUAAGCAUCGAUAGCUGCGAAGCGAAGCAU .......................((((.((((((...((((....(((((((((..(((.....)))......))))))))).....))))...).)))))....)))).. (-14.81 = -16.67 + 1.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:02 2011