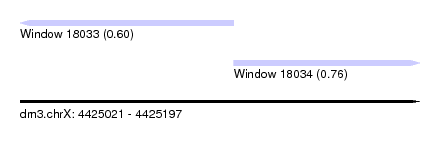
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,425,021 – 4,425,197 |
| Length | 176 |
| Max. P | 0.759749 |

| Location | 4,425,021 – 4,425,115 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Shannon entropy | 0.40960 |
| G+C content | 0.51708 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.35 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
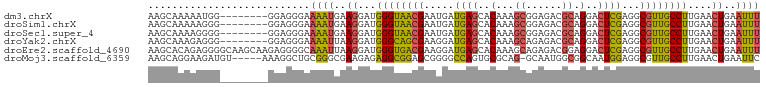
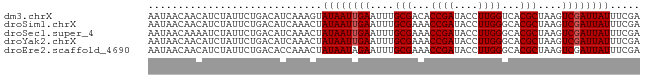
>dm3.chrX 4425021 94 - 22422827 AAGCAAAAAUGG--------GGAGGGAAAAUGAAGGAUGGGUAACGAAUGAUGAGCACAAAGCGGAGACGCAGGACUCGAGGCGUUGCCUUGAACUGAAUUU ............--------.......((((..((...((((((((.....((((..(...(((....))).)..))))...))))))))....))..)))) ( -23.60, z-score = -2.72, R) >droSim1.chrX 3424659 94 - 17042790 AAGCAAAAAGGG--------GGAGGGAAAAUGAAGGAUGGGUAACGAAUGAUGAGCACAAAGCGGAGACGCAGGACUCGAGGCGUUGCCUUGAACUGAAUUU ............--------.......((((..((...((((((((.....((((..(...(((....))).)..))))...))))))))....))..)))) ( -23.60, z-score = -2.69, R) >droSec1.super_4 3876761 94 + 6179234 AAGCAAAAGGGG--------GGAGGGAAAAUGAAGGAUGGGUAACGAAUGAUGAGCACAAAGCGGAGACGCAGGACUCGAGGCGUUGCCUUGAACUGAAUUU ............--------.......((((..((...((((((((.....((((..(...(((....))).)..))))...))))))))....))..)))) ( -23.60, z-score = -2.62, R) >droYak2.chrX 17758167 94 + 21770863 AAGCAAAGAGGG--------GGAGGGAAAAUUAAGGAUGGGCAGCGAAGGAUGAGCACAAAGCAGAGACGCAGGACUCGAGGCGUUGCCUUGAACUGAAUUU ............--------.......(((((.((...((((((((.....((((..(...((......)).)..))))...))))))))....)).))))) ( -19.40, z-score = -0.45, R) >droEre2.scaffold_4690 1803618 102 - 18748788 AAGCACAGAGGGGCAAGCAAGAGGGGCAAAUUAAGGAUGGGUGACGAAGGAUGAGCACAAAGCAGAGACGGAGGACUCGAGGCGUUGCCUUGAACUGAAUUU .....(((.(((((((((..(((................(....).........((.....))............)))...)).)))))))...)))..... ( -21.80, z-score = -0.67, R) >droMoj3.scaffold_6359 4056644 96 - 4525533 AAGCAGGAAGAUGU-----AAAGGCUGCGGGCGAAGAGAGGCGGAGCGGGGCCAGUGCGCAG-GCAAUGGCGGCAAUGGAGGCGUUGCCUUGAACUGAAUUC ...(((........-----.....((((..((........))...)))).((((.(((....-))).))))(((((((....))))))).....)))..... ( -29.00, z-score = 0.12, R) >consensus AAGCAAAAAGGG________GGAGGGAAAAUGAAGGAUGGGUAACGAAGGAUGAGCACAAAGCGGAGACGCAGGACUCGAGGCGUUGCCUUGAACUGAAUUU ...........................((((..((...((((((((.....((((..(...((......)).)..))))...))))))))....))..)))) (-13.68 = -13.35 + -0.33)
| Location | 4,425,115 – 4,425,197 |
|---|---|
| Length | 82 |
| Sequences | 5 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 97.07 |
| Shannon entropy | 0.05282 |
| G+C content | 0.35366 |
| Mean single sequence MFE | -13.26 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
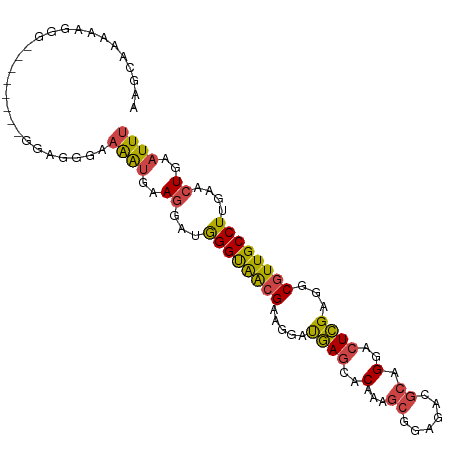
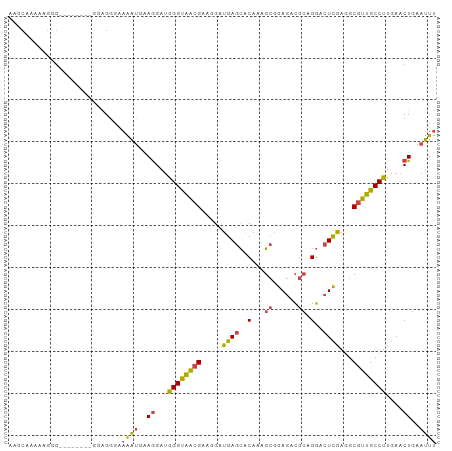
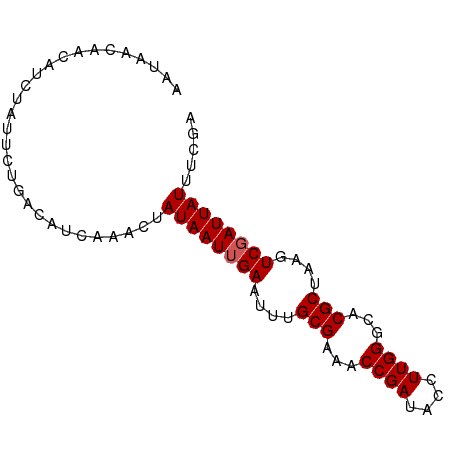
>dm3.chrX 4425115 82 + 22422827 AAUAACAACAUCUAUUCUGACAUCAAAGUAUAAUUGAAUUUGCGACACCGAUACCUUGGUCACGCUAAGUCGAUUAUUUCGA ...........................(.((((((((....(((..(((((....)))))..)))....))))))))..).. ( -14.20, z-score = -1.26, R) >droSim1.chrX 3424753 82 + 17042790 AAUAACAACAUCUAUUCUGACAUCAAACUAUAAUUGAAUUUGCGAAACCGAUACCUUGGGCACGCUAAGUCGAUUAUUUCGA .............................((((((((....(((...((((....))))...)))....))))))))..... ( -13.70, z-score = -1.62, R) >droSec1.super_4 3876855 82 - 6179234 AAUAACAAAAUCUAUUCUGACAUCAAACUAUAAUUGAAUUUGCGAAACCGAUACCUUGGGCACGCUAAGUCGAUUAUUUCGA .............................((((((((....(((...((((....))))...)))....))))))))..... ( -13.70, z-score = -1.60, R) >droYak2.chrX 17758261 82 - 21770863 AAUAACAACAUCUAUUCUGACAUCAAACUAUAAUUGAAUUUGCGAAACCGAUACCUUGGGCACGCUAAGUCGAUUAUUUCGA .............................((((((((....(((...((((....))))...)))....))))))))..... ( -13.70, z-score = -1.62, R) >droEre2.scaffold_4690 1803720 82 + 18748788 AAUAACAACAUCUAUUCUGACACCAAACUAUAAUAGAAUUUGCGAAACCGAUACCUUGGGCACGCUAAGUCGAUUAUUUCGA ..........((((((.((.........)).))))))....(((...((((....))))...)))....((((.....)))) ( -11.00, z-score = -0.72, R) >consensus AAUAACAACAUCUAUUCUGACAUCAAACUAUAAUUGAAUUUGCGAAACCGAUACCUUGGGCACGCUAAGUCGAUUAUUUCGA .............................((((((((....(((...((((....))))...)))....))))))))..... (-12.74 = -12.94 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:15:01 2011