
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,420,900 – 4,420,988 |
| Length | 88 |
| Max. P | 0.886270 |

| Location | 4,420,900 – 4,420,988 |
|---|---|
| Length | 88 |
| Sequences | 5 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 64.23 |
| Shannon entropy | 0.60747 |
| G+C content | 0.42227 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -11.21 |
| Energy contribution | -10.89 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797946 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
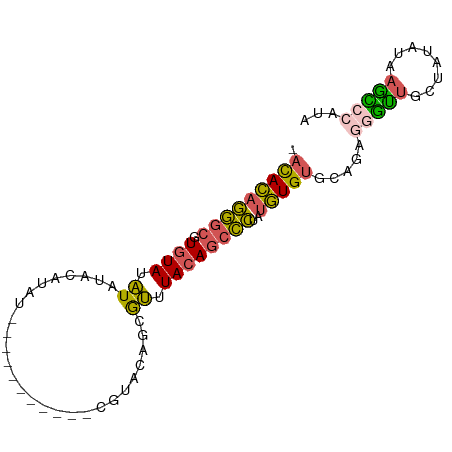
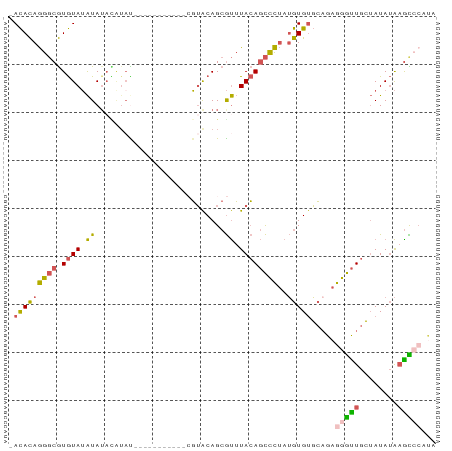
>dm3.chrX 4420900 88 + 22422827 -ACACAGGGCGUGUAUAUAUAUAUACGUAACGUACAGCGUACAGCGUUUACAGCCCUAUGUGUGCCGAGGGUUGCUAUAUAAGCCCAUG -(((((((((((((((....)))))).....(((.((((.....))))))).))))..))))).....(((((........)))))... ( -28.80, z-score = -1.17, R) >droSim1.chrX 3420623 77 + 17042790 -ACACAGGGCGUGUAUAUAUACGUAU-----------CGUACAGCGUUUACAGCCCUAUGUGUGCAGAGGGUUGCUAUAUAAGCCCAUA -.....((((.((((((((.((((..-----------......)))).))(((((((.((....)).))))))).)))))).))))... ( -26.40, z-score = -1.84, R) >droSec1.super_4 3872684 77 - 6179234 -ACACAGGGCGUGUAUAUAUACGUAU-----------CGUACAGCGUUUACAGCCCUAUGUGUGCAGAGGGUUGCUAUAUAAGCCCAUA -.....((((.((((((((.((((..-----------......)))).))(((((((.((....)).))))))).)))))).))))... ( -26.40, z-score = -1.84, R) >droYak2.chrX 17753240 68 - 21770863 -ACACAGGGCGUGUAUAUA------C------------GUACAGCGUUUACAGCCCUAUGUGUGCAGAGGGUUGCUAUAUACAUAAG-- -.....((((.((((...(------(------------(.....))).))))))))(((((((((((....))))...)))))))..-- ( -20.40, z-score = -1.04, R) >droGri2.scaffold_15252 15196261 82 + 17193109 AAUAUAAAUCAUAUAAGUAUAUAUAUCUAG-----AGAUGGCAGCACAUAAAAUUUAAAAUGCUGAGCAAACUAA-AUA-GAGUGCACG ................((((...(((.(((-----......(((((..............)))))......))).-)))-..))))... ( -9.64, z-score = 0.44, R) >consensus _ACACAGGGCGUGUAUAUAUACAUAU___________CGUACAGCGUUUACAGCCCUAUGUGUGCAGAGGGUUGCUAUAUAAGCCCAUA .(((((((((.((((.((...........................)).))))))))..))))).....(((((........)))))... (-11.21 = -10.89 + -0.32)

| Location | 4,420,900 – 4,420,988 |
|---|---|
| Length | 88 |
| Sequences | 5 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 64.23 |
| Shannon entropy | 0.60747 |
| G+C content | 0.42227 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.37 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886270 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
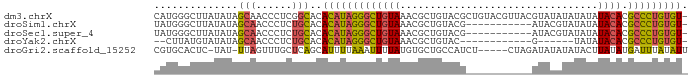
>dm3.chrX 4420900 88 - 22422827 CAUGGGCUUAUAUAGCAACCCUCGGCACACAUAGGGCUGUAAACGCUGUACGCUGUACGUUACGUAUAUAUAUAUACACGCCCUGUGU- .....(((.....)))............(((((((((((((...((.....))((((((...))))))......)))).)))))))))- ( -24.40, z-score = -0.05, R) >droSim1.chrX 3420623 77 - 17042790 UAUGGGCUUAUAUAGCAACCCUCUGCACACAUAGGGCUGUAAACGCUGUACG-----------AUACGUAUAUAUACACGCCCUGUGU- ..............(((......)))..(((((((((((((.....((((((-----------...))))))..)))).)))))))))- ( -22.50, z-score = -1.20, R) >droSec1.super_4 3872684 77 + 6179234 UAUGGGCUUAUAUAGCAACCCUCUGCACACAUAGGGCUGUAAACGCUGUACG-----------AUACGUAUAUAUACACGCCCUGUGU- ..............(((......)))..(((((((((((((.....((((((-----------...))))))..)))).)))))))))- ( -22.50, z-score = -1.20, R) >droYak2.chrX 17753240 68 + 21770863 --CUUAUGUAUAUAGCAACCCUCUGCACACAUAGGGCUGUAAACGCUGUAC------------G------UAUAUACACGCCCUGUGU- --............(((......)))..(((((((((((((.(((.....)------------)------)...)))).)))))))))- ( -21.00, z-score = -2.07, R) >droGri2.scaffold_15252 15196261 82 - 17193109 CGUGCACUC-UAU-UUAGUUUGCUCAGCAUUUUAAAUUUUAUGUGCUGCCAUCU-----CUAGAUAUAUAUACUUAUAUGAUUUAUAUU ...((((..-.((-((((..(((...)))..)))))).....))))........-----.((((((((((....))))).))))).... ( -12.60, z-score = -0.38, R) >consensus CAUGGGCUUAUAUAGCAACCCUCUGCACACAUAGGGCUGUAAACGCUGUACG___________AUACAUAUAUAUACACGCCCUGUGU_ ..............(((......)))..(((((((((((((.................................)))).))))))))). (-11.77 = -11.37 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:57 2011