| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,405,198 – 4,405,304 |
| Length | 106 |
| Max. P | 0.742022 |

| Location | 4,405,198 – 4,405,304 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Shannon entropy | 0.47554 |
| G+C content | 0.61756 |
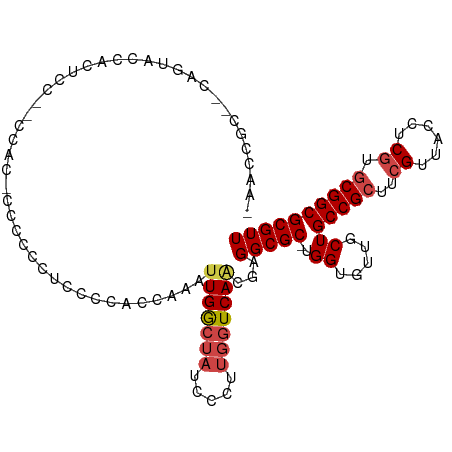
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -17.43 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
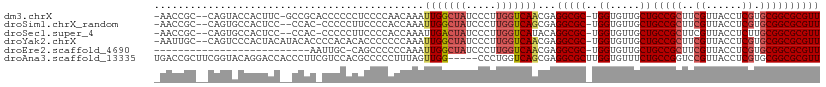
>dm3.chrX 4405198 106 - 22422827 -AACCGC--CAGUACCACUUC-GCCGCACCCCCCUCCCCAACAAAUUGGCUAUCCCUUGGUCAACGAGGCGC-UGGUGUUGCUGCCGCUUCGUUACCUCGUGCGGCGCGUU -...(((--.((.....))..-(((((((........((((....)))).........(((.(((((((((.-.((.....))..))))))))))))..)))))))))).. ( -36.30, z-score = -2.03, R) >droSim1.chrX_random 1595312 104 - 5698898 -AACCGC--CAGUGCCACUCC--CCAC-CCCCCUUCCCCACCAAAUUGGCUAUCCCUUGGUCAGCGAGGCGC-UGGUGUUGCUGCCGCUUCGUUACCUCGUGCGGCGCGUU -...(((--(((((((.....--....-..........((......))(((..((...))..)))..)))))-)))))..((.(((((..((......)).)))))))... ( -35.90, z-score = -2.07, R) >droSec1.super_4 3857220 104 + 6179234 -AACCGC--CAGUGCCACUCC--CCAC-CCCCCUUCCCCACCAAAUUGACUAUCCCUUGGUCAUACAGGCGC-UGGUGUUGCUGCCGCUUCGUUACCUCUUGCGGCGCGUU -...(((--(((((((.....--....-.....((........)).((((((.....))))))....)))))-)))))..((.(((((.............)))))))... ( -32.92, z-score = -3.41, R) >droYak2.chrX 17737731 107 + 21770863 -AAUUGC--CAGUCCCACUACAUACACCCCACACACCCCCCCAAAUUGGCUAUCCCUUGGUCAACGAGGCGC-UGGUGUUGCUGCCGCUUCGUUACCUCGUGCGGCGCGUU -......--.......................(((((..(((...(((((((.....)))))))...)).).-.))))).((.(((((..((......)).)))))))... ( -27.70, z-score = 0.02, R) >droEre2.scaffold_4690 1782023 83 - 18748788 --------------------------AAUUGC-CAGCCCCCCAAAUUGGCUAUCCCUUGGUCAACGAGGCGC-UGGUGUUGCUGCCGCUUCGUUACCUCGUGCGGCGCGUU --------------------------....((-((((.((.(...(((((((.....))))))).).)).))-))))...((.(((((..((......)).)))))))... ( -32.00, z-score = -1.95, R) >droAna3.scaffold_13335 3295967 106 + 3335858 UGACCGCUUCGGUACAGGACCACCCUUCGUCCACGCCCCCUUUAGUUGG-----CCCUGGUCAGCGAGGCGCUUGGUGUUUCUGCCGGUCCGUUACCUCGUGCGGCGCGUU .(.((((...((((..(((((............((((.......(((((-----(....))))))..))))...(((......))))))))..))))....)))))..... ( -37.60, z-score = 0.01, R) >consensus _AACCGC__CAGUACCACUCC__CCAC_CCCCCCUCCCCACCAAAUUGGCUAUCCCUUGGUCAACGAGGCGC_UGGUGUUGCUGCCGCUUCGUUACCUCGUGCGGCGCGUU ...((((.......................................((((((.....))))))((((((((..((((......))))...))...))))))))))...... (-17.43 = -18.13 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:55 2011