
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,404,391 – 4,404,481 |
| Length | 90 |
| Max. P | 0.704871 |

| Location | 4,404,391 – 4,404,481 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.00 |
| Shannon entropy | 0.49188 |
| G+C content | 0.41777 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -12.77 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4404391 90 + 22422827 CAAUUUCAAAUCCAAUCGAAUGCAAUUCUAAUAACCAAAUCCGGGGACGUAUGCAAUUUGCAUGGCCAGC---UGCUGGCCAAAAGUUGAAAC----------------- ...((((((..........((((((.....(((.((.......))....))).....))))))((((((.---..)))))).....)))))).----------------- ( -21.00, z-score = -1.03, R) >droSim1.chrX_random 1594537 90 + 5698898 CAAUUUCAAAUCCAAUCCAAUGCAAUUCUAAUAACCAAAUCCGGUGCCGUAUGCAAUUUGCAUGGCCAGC---UGCUGGCCAAAAGUUGAAGC----------------- ...((((((..........((((((........(((......)))((.....))...))))))((((((.---..)))))).....)))))).----------------- ( -21.50, z-score = -1.12, R) >droSec1.super_4 3856448 90 - 6179234 CAAUUUCAAAUCCAAUCCAAUGCAAUUCUAAUAACCAAAUCCGGUGCCGUAUGCAAUUUGCAUGGCCAGC---UGCUGGCCAAAAGUUGAAGC----------------- ...((((((..........((((((........(((......)))((.....))...))))))((((((.---..)))))).....)))))).----------------- ( -21.50, z-score = -1.12, R) >droYak2.chrX 17736879 90 - 21770863 CAAUUUCAAAUCCAAUCCAAUGCAAUUCUAAUAACCAAAUCCGGGGCCGUAUGCAAUUUGCAUGGCCAGC---UGCUGGCCAAAAGUUGAAAC----------------- ...((((((.......(((..((...........((......))((((..((((.....)))))))).))---...))).......)))))).----------------- ( -21.24, z-score = -0.78, R) >droEre2.scaffold_4690 1781190 90 + 18748788 CAAUUUCAAAUCCAAUCCAAUGCAAUUCUAAUAACCAAAUCCGGGGCCGUAUGCAAUUUGCAUGGCCAGC---UGCUGGCCAAAAGUUGAAAC----------------- ...((((((.......(((..((...........((......))((((..((((.....)))))))).))---...))).......)))))).----------------- ( -21.24, z-score = -0.78, R) >droGri2.scaffold_15203 5909065 87 - 11997470 CAAUUUCAAAUCCAAUUCUAUGGAAUUCUAAUAAACAAAUCCAACAGC--AUGCAAAUUUCAUGGCCAGA---UGCUGGCCAA-AACGGAAAA----------------- ..........((((......))))...............(((....(.--...)........(((((((.---..))))))).-...)))...----------------- ( -16.90, z-score = -1.58, R) >droVir3.scaffold_12928 5499793 88 + 7717345 CAGUUUCAAAUCCAAUUCAAUGGCAUUCUAAUAACCAAAUCCUACUAC--AUGCAAUUUUCAUGGCCAGC---UGCUGGCCAACAACCGAAAA----------------- ...((((...............((((......................--))))........(((((((.---..)))))))......)))).----------------- ( -14.35, z-score = -0.88, R) >droMoj3.scaffold_6359 4019614 87 + 4525533 CAAUUUCAAAUCCAAUUCUAUGGUAUUCUAAUAACCAAAUCCAUCUGC--AUGCAAUUUUCAUGGCCAGU---UGCUGGCCAA-AACUGAAAA----------------- ...(((((............((((.........))))...........--............(((((((.---..))))))).-...))))).----------------- ( -16.20, z-score = -1.35, R) >droWil1.scaffold_181150 3029726 84 - 4952429 UAGUUUCAAAUCCAAUUCAAUGCAAUUCGAAUAACCAAUUCCAA-ACUGCAUGCAAUAUGAAAGACCGCCUG-CACAGGCUUGCCC------------------------ ...................(((((....((((.....))))...-..)))))((((..(....)...((((.-...))))))))..------------------------ ( -11.40, z-score = 0.01, R) >droAna3.scaffold_13335 3294892 99 - 3335858 CAAUUUCAAAUCCAAUCCAAUGCAAUUCUAAUAACCAAAUCCGGCCACU-AUGCAAUUUGCAUGCACAGC---UGCUGGCCAAAAACUUUCAACCAACAACAC------- ..........................................(((((..-((((.....))))(((....---))))))))......................------- ( -14.10, z-score = -1.04, R) >dp4.chrXL_group1a 5515001 109 - 9151740 CAAUUUCAAAUCCAAUUCAAUGCAAUUCUAAUAACCAAAUCC-AUGCCGUAUGCAAUUUGCAUGGCCAGCAGCUGCUGGCCAAACUGUGAGCCACAUGGGCAGGGUCUCU ......................................((((-.((((.((((...((..((((((((((....))))))))...))..))...)))))))))))).... ( -31.30, z-score = -1.06, R) >droPer1.super_11 2650374 109 - 2846995 CAAUUUCAAAUCCAAUUCAAUGCAAUUCUAAUAACCAAAUCC-AUGCCGUAUGCAAUUUGCAUGGCCAGCAGCUGCUGGCCAAACUGUGAGCCGCAUGGGCAGGGUCUCU ......................................((((-.((((.(((((..((..((((((((((....))))))))...))..))..))))))))))))).... ( -35.30, z-score = -2.02, R) >consensus CAAUUUCAAAUCCAAUCCAAUGCAAUUCUAAUAACCAAAUCCGGCGCCGUAUGCAAUUUGCAUGGCCAGC___UGCUGGCCAAAAGUUGAAAC_________________ ....................(((((................................)))))((((((((....))))))))............................ (-12.77 = -13.02 + 0.25)

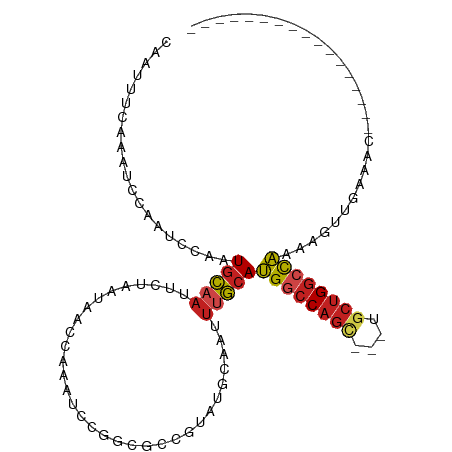

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:54 2011