
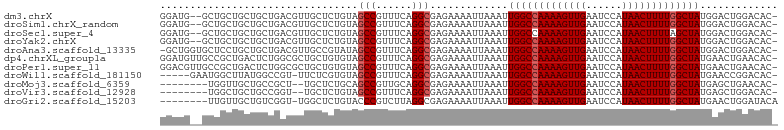
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,399,882 – 4,399,982 |
| Length | 100 |
| Max. P | 0.888418 |

| Location | 4,399,882 – 4,399,982 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Shannon entropy | 0.31208 |
| G+C content | 0.46035 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4399882 100 + 22422827 GGAUG--GCUGCUGCUGCUGACGUUGCUCUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGGACUGGACAC- .((((--(((((.((.((....)).))...)))))))))((((...............(((((((((((((......)))))))))))))....))))....- ( -32.01, z-score = -2.48, R) >droSim1.chrX_random 1589543 100 + 5698898 GGAUG--GCUGCUGCUGCUGACGUUGCUCUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGGACUGGACAC- .((((--(((((.((.((....)).))...)))))))))((((...............(((((((((((((......)))))))))))))....))))....- ( -32.01, z-score = -2.48, R) >droSec1.super_4 3851784 100 - 6179234 GGAUG--GCUGCUGCUGCUGACGUUGCUCUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUAGCUAUGGACUGGACAC- (((((--(((((.((.((....)).))...))))))))))..(((.((.........)).)))....(((.(.((((((.(.....).)))))).).)))..- ( -28.70, z-score = -1.54, R) >droYak2.chrX 17732322 100 - 21770863 GGAUG--GCUGCUGCUGCUGACGUUGCUCUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGGACUGGACAC- .((((--(((((.((.((....)).))...)))))))))((((...............(((((((((((((......)))))))))))))....))))....- ( -32.01, z-score = -2.48, R) >droAna3.scaffold_13335 3289624 101 - 3335858 -GCUGGUGCUCCUGCUGCUGACGUUGCCGUAUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGGACUGGACAC- -.(..((.(((.((((...((((..((......))))))...)))).)).........(((((((((((((......))))))))))))).).))..)....- ( -29.90, z-score = -1.55, R) >dp4.chrXL_group1a 5508407 102 - 9151740 GGAUGUUGCCGCUGACUCUGGCGCUGCUGUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGAACUGAACAC- ...(.((((((((......)))(((((...))))).......))))).).........(((((((((((((......)))))))))))))............- ( -30.40, z-score = -2.10, R) >droPer1.super_11 2643710 102 - 2846995 GGACGUUGCCGCUGACUCUGGCGCUGCUGUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGAACUGAACAC- .....((((((((......)))(((((...))))).......)))))...........(((((((((((((......)))))))))))))............- ( -29.90, z-score = -1.80, R) >droWil1.scaffold_181150 3025335 96 - 4952429 -----GAAUGGCUUAUGGCCGU-UUCUCGUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGAACCGGACAC- -----(((((((.....)))))-))...((((.(((......))).............(((((((((((((......)))))))))))))........))))- ( -31.20, z-score = -3.13, R) >droMoj3.scaffold_6359 4014729 92 + 4525533 --------UGGUUGCUGCCGCU--UGCUCUGCAGCCGUUGCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGAGCUGAACAC- --------..((((((....((--(((.((((((...)))))))))))..........(((((((((((((......)))))))))))))..)))..)))..- ( -31.60, z-score = -2.55, R) >droVir3.scaffold_12928 5495819 92 + 7717345 --------UGGCUGCUGCCGGU--UGCUCUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGAGCUGGACAC- --------.(((....))).((--(((((....(((......))).............(((((((((((((......))))))))))))).))))..)))..- ( -32.10, z-score = -2.76, R) >droGri2.scaffold_15203 5904525 94 - 11997470 --------UUGUUGCUGUCGGU-UGGCUCUGUACCCGUCUUAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGAACUGGAUACA --------((.(((((...((.-(((........))).))..))))).))........(((((((((((((......)))))))))))))............. ( -23.50, z-score = -1.10, R) >consensus GGAUG__GCUGCUGCUGCUGACGUUGCUCUGUAGCCGUUUCAGGCGAGAAAAUUAAAUUGGCCAAAAGUUGAAUCCAUAACUUUUGGCUAUGAACUGGACAC_ .................................(((......))).............(((((((((((((......)))))))))))))............. (-19.70 = -19.88 + 0.18)


| Location | 4,399,882 – 4,399,982 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.86 |
| Shannon entropy | 0.31208 |
| G+C content | 0.46035 |
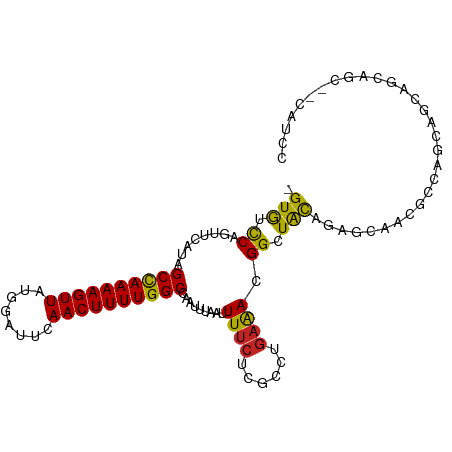
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -16.50 |
| Energy contribution | -16.07 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4399882 100 - 22422827 -GUGUCCAGUCCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACAGAGCAACGUCAGCAGCAGCAGC--CAUCC -..((..((((((((((.....))))))))))..))...((((..........((((.(((......)))...))))........((....)).))--))... ( -23.10, z-score = -1.22, R) >droSim1.chrX_random 1589543 100 - 5698898 -GUGUCCAGUCCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACAGAGCAACGUCAGCAGCAGCAGC--CAUCC -..((..((((((((((.....))))))))))..))...((((..........((((.(((......)))...))))........((....)).))--))... ( -23.10, z-score = -1.22, R) >droSec1.super_4 3851784 100 + 6179234 -GUGUCCAGUCCAUAGCUAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACAGAGCAACGUCAGCAGCAGCAGC--CAUCC -..((..((((((((((.....))))))))))..))...((((..........((((.(((......)))...))))........((....)).))--))... ( -23.80, z-score = -1.34, R) >droYak2.chrX 17732322 100 + 21770863 -GUGUCCAGUCCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACAGAGCAACGUCAGCAGCAGCAGC--CAUCC -..((..((((((((((.....))))))))))..))...((((..........((((.(((......)))...))))........((....)).))--))... ( -23.10, z-score = -1.22, R) >droAna3.scaffold_13335 3289624 101 + 3335858 -GUGUCCAGUCCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUAUACGGCAACGUCAGCAGCAGGAGCACCAGC- -((((..........((((((((((........))))))))))................((((.....(((..(((....))).)))..)))).))))....- ( -28.30, z-score = -2.00, R) >dp4.chrXL_group1a 5508407 102 + 9151740 -GUGUUCAGUUCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACACAGCAGCGCCAGAGUCAGCGGCAACAUCC -((((..........((((((((((........))))))))))...............(((......))).)))).((.((..........)).))....... ( -24.00, z-score = -0.98, R) >droPer1.super_11 2643710 102 + 2846995 -GUGUUCAGUUCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACACAGCAGCGCCAGAGUCAGCGGCAACGUCC -..(((.((((((((((.....)))))))))).)))((((((((........((((......))))..(((....))).).)))))))...(((....))).. ( -25.70, z-score = -1.26, R) >droWil1.scaffold_181150 3025335 96 + 4952429 -GUGUCCGGUUCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACACGAGAA-ACGGCCAUAAGCCAUUC----- -(((.(((.......((((((((((........)))))))))).........(((((((((......))).....)))))-)))).))).........----- ( -26.20, z-score = -2.44, R) >droMoj3.scaffold_6359 4014729 92 - 4525533 -GUGUUCAGCUCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGCAACGGCUGCAGAGCA--AGCGGCAGCAACCA-------- -.(((((.(((....((((((((((........))))))))))...............(((((((.....))))).)).--))).).))))....-------- ( -26.60, z-score = -1.27, R) >droVir3.scaffold_12928 5495819 92 - 7717345 -GUGUCCAGCUCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACAGAGCA--ACCGGCAGCAGCCA-------- -.......((((...((((((((((........))))))))))...............(((......)))....)))).--...(((....))).-------- ( -28.30, z-score = -2.70, R) >droGri2.scaffold_15203 5904525 94 + 11997470 UGUAUCCAGUUCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUAAGACGGGUACAGAGCCA-ACCGACAGCAACAA-------- (((((((........((((((((((........))))))))))...........(((......))).)))))))......-..............-------- ( -21.40, z-score = -1.93, R) >consensus _GUGUCCAGUUCAUAGCCAAAAGUUAUGGAUUCAACUUUUGGCCAAUUUAAUUUUCUCGCCUGAAACGGCUACAGAGCAACGCCAGCAGCAGCAGC__CAUCC .(((.((........((((((((((........)))))))))).........((((......)))).)).))).............................. (-16.50 = -16.07 + -0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:53 2011