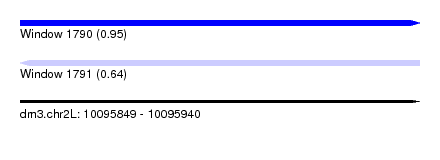
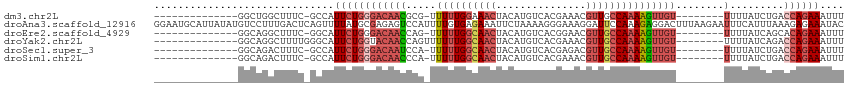
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,095,849 – 10,095,940 |
| Length | 91 |
| Max. P | 0.947734 |

| Location | 10,095,849 – 10,095,940 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.93 |
| Shannon entropy | 0.48370 |
| G+C content | 0.40706 |
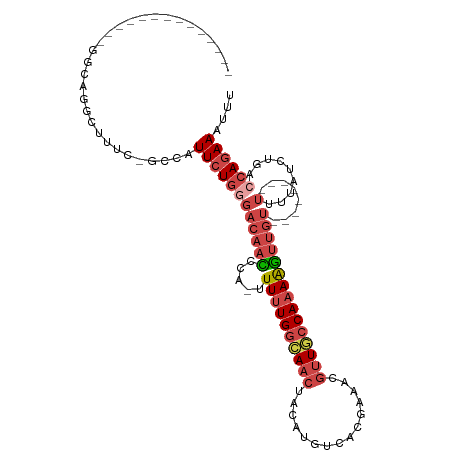
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -11.10 |
| Energy contribution | -12.44 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
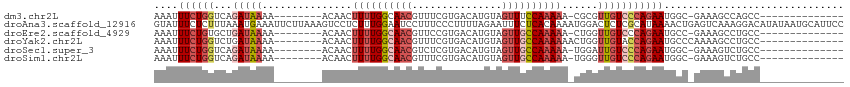
>dm3.chr2L 10095849 91 + 23011544 AAAUUUCUGGUCAGAUAAAA--------ACAACUUUUGGCAACGUUUCGUGACAUGUAGUUUCCAAAAA-CGCGUUGUCCCAGAAUGGC-GAAAGCCAGCC-------------- ....((((((...(((((..--------.....((((((.(((....(((...)))..))).)))))).-....)))))))))))((((-....))))...-------------- ( -24.10, z-score = -1.97, R) >droAna3.scaffold_12916 13928493 115 + 16180835 GUAUUUCUCUUUAAAUGAAAUUCUUAAAGUCCUCUUUGGAAUCCUUUCCCUUUUAGAAUUUCUCACAAAAUGGACUCUCGCAUAAAACUGAGUCAAAGGACAUAUAAUGCAUUCC (((((..(((((....((((((((.((((........((((....)))))))).))))))))..........(((((............))))).))))).....)))))..... ( -22.20, z-score = -2.27, R) >droEre2.scaffold_4929 10711251 91 + 26641161 AAAUUUCUGUGCUGAUAAAA--------ACAACUUUUGGCAACGUUCCGUGACAUGUAGUUGCCAAAAA-CUGGUUGUCCCAGAAUGCC-GAAAGCCUGCC-------------- ....................--------....(((((((((...(((.(.((((..(((((......))-)))..)))).).)))))))-)))))......-------------- ( -21.40, z-score = -1.51, R) >droYak2.chr2L 12779781 93 + 22324452 AAAUUUCUGGUCUGAUAAAA--------ACAACUUUUGGCAACGUUUCGUGACAUGUAGUUGCCAAAAAACUGGUUGUACCAGAAUGCCCAAAAGCCUGCC-------------- ....(((((((.........--------(((((((((((((((....(((...)))..))))))))).....)))))))))))))................-------------- ( -23.20, z-score = -1.84, R) >droSec1.super_3 5533831 91 + 7220098 AAAUUUCUGGUCAGAUAAAA--------ACAACUUUUGGCAACGUCUCGUGACAUGUAGUUGCCAAAAA-UGGAUUGUCCCAGAAUGGC-GAAAGUCUGCC-------------- ....((((((...(((((..--------.((..(((((((((((((....))).....)))))))))).-))..))))))))))).(((-....)))....-------------- ( -28.60, z-score = -3.52, R) >droSim1.chr2L 9884396 91 + 22036055 AAAUUUCUGGUCAGAUAAAA--------ACAACUUUUGGCAACGUUUCGUGACAUGUAGUUGCCAAAAA-UGGGUUGUCCCAGAAUGGC-GAAAGUCUGCC-------------- ....((((((...(((((..--------.((..((((((((((....(((...)))..)))))))))).-))..))))))))))).(((-....)))....-------------- ( -28.10, z-score = -2.94, R) >consensus AAAUUUCUGGUCAGAUAAAA________ACAACUUUUGGCAACGUUUCGUGACAUGUAGUUGCCAAAAA_CGGGUUGUCCCAGAAUGCC_GAAAGCCUGCC______________ ....((((((..................(((((((((((((((...............))))))))).....)))))).)))))).............................. (-11.10 = -12.44 + 1.34)
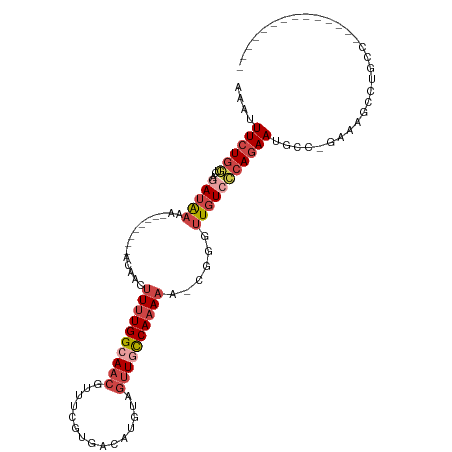
| Location | 10,095,849 – 10,095,940 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.93 |
| Shannon entropy | 0.48370 |
| G+C content | 0.40706 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -11.09 |
| Energy contribution | -12.54 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10095849 91 - 23011544 --------------GGCUGGCUUUC-GCCAUUCUGGGACAACGCG-UUUUUGGAAACUACAUGUCACGAAACGUUGCCAAAAGUUGU--------UUUUAUCUGACCAGAAAUUU --------------...((((....-))))((((((((((((...-.((((((.(((...............))).)))))))))))--------).........)))))).... ( -20.06, z-score = -0.14, R) >droAna3.scaffold_12916 13928493 115 - 16180835 GGAAUGCAUUAUAUGUCCUUUGACUCAGUUUUAUGCGAGAGUCCAUUUUGUGAGAAAUUCUAAAAGGGAAAGGAUUCCAAAGAGGACUUUAAGAAUUUCAUUUAAAGAGAAAUAC (((((...(((...((((((((((((.((.....))..)))))..(((((.((....)).))))).((((....))))..)))))))..)))..)))))................ ( -24.00, z-score = -0.74, R) >droEre2.scaffold_4929 10711251 91 - 26641161 --------------GGCAGGCUUUC-GGCAUUCUGGGACAACCAG-UUUUUGGCAACUACAUGUCACGGAACGUUGCCAAAAGUUGU--------UUUUAUCAGCACAGAAAUUU --------------(((((..((((-(.....((((.....))))-....(((((......))))))))))..)))))....((((.--------......)))).......... ( -23.00, z-score = -1.26, R) >droYak2.chr2L 12779781 93 - 22324452 --------------GGCAGGCUUUUGGGCAUUCUGGUACAACCAGUUUUUUGGCAACUACAUGUCACGAAACGUUGCCAAAAGUUGU--------UUUUAUCAGACCAGAAAUUU --------------.....(((....))).((((((((((((.....((((((((((...............)))))))))))))))--------.........))))))).... ( -24.06, z-score = -1.50, R) >droSec1.super_3 5533831 91 - 7220098 --------------GGCAGACUUUC-GCCAUUCUGGGACAAUCCA-UUUUUGGCAACUACAUGUCACGAGACGUUGCCAAAAGUUGU--------UUUUAUCUGACCAGAAAUUU --------------(((........-))).((((((((((((...-.((((((((((.....(((....))))))))))))))))))--------).........)))))).... ( -26.90, z-score = -3.23, R) >droSim1.chr2L 9884396 91 - 22036055 --------------GGCAGACUUUC-GCCAUUCUGGGACAACCCA-UUUUUGGCAACUACAUGUCACGAAACGUUGCCAAAAGUUGU--------UUUUAUCUGACCAGAAAUUU --------------(((........-))).((((((((((((...-.((((((((((...............)))))))))))))))--------).........)))))).... ( -26.16, z-score = -3.17, R) >consensus ______________GGCAGGCUUUC_GCCAUUCUGGGACAACCCA_UUUUUGGCAACUACAUGUCACGAAACGUUGCCAAAAGUUGU________UUUUAUCUGACCAGAAAUUU ..............................((((((.(((((.....((((((((((...............)))))))))))))))..................)))))).... (-11.09 = -12.54 + 1.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:33 2011