| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,384,509 – 4,384,599 |
| Length | 90 |
| Max. P | 0.948826 |

| Location | 4,384,509 – 4,384,599 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.69 |
| Shannon entropy | 0.47191 |
| G+C content | 0.52431 |
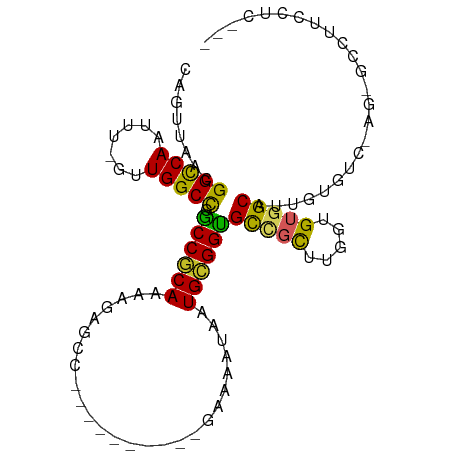
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -20.00 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
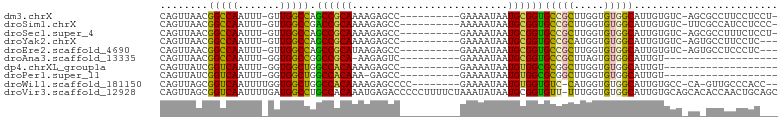
>dm3.chrX 4384509 90 + 22422827 CAGUUAACGGCCAAUUU-GUUGGCCAGCCGCAAAAGAGCC----------GAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUC-AGCGCCUUCCUCCU- ..(((.((((((((...-.)))))).((((((.(((.(((----------(.........))))....)))..)))))).....)).-)))...........- ( -29.60, z-score = -0.63, R) >droSim1.chrX 3390628 90 + 17042790 CAGUUAACGGCCAAUUU-GUUGGCCGACCGCAAAAGAGCC----------AAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUC-UUCGCCAUCCUCCC- ..((...(((((((...-.)))))))...))....(((..----------.......((((((((((((.....)))))))))))).-.........)))..- ( -29.21, z-score = -1.48, R) >droSec1.super_4 3836844 90 - 6179234 CAGUUAACGGCCAAUUU-GUUGGCCAGCCGCAAAAGAGCC----------GAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUC-AGCGCCUUUCUCCU- ..(((.((((((((...-.)))))).((((((.(((.(((----------(.........))))....)))..)))))).....)).-)))...........- ( -29.60, z-score = -0.65, R) >droYak2.chrX 17717063 88 - 21770863 CAGUUAACGGCCAAUUU-GUUGGCCAGCCGCAAAAGAGCC----------GAAAAUAAUGCGGUGCCGCAUGGUGUGGCAUUGUGUC-AGUGCCUUCCUC--- .......((((...(((-((.((....)))))))...)))----------)......(((((....)))))((...(((((((...)-))))))..))..--- ( -29.70, z-score = -1.19, R) >droEre2.scaffold_4690 1760992 88 + 18748788 CAGUUAACGGCCAAUUU-GUUGGCCAGCCGCAUAAGAGCC----------GAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUC-AGUGCCUCCCUC--- ........((((((...-.)))))).(((((((.......----------.......))))))).......((.(.(((((((...)-)))))).)))..--- ( -30.44, z-score = -1.37, R) >droAna3.scaffold_13335 3272359 72 - 3335858 CAGUUAACGGCCAAUUU-GGUGGCCGGCCGCA-AAGAGUC----------GAAAAUAAUGCGGUGCCGCUUAGUGUGGCAUUGU------------------- .......((((((....-..))))))((((((-.......----------........))))))(((((.....))))).....------------------- ( -27.56, z-score = -2.17, R) >dp4.chrXL_group1a 5491688 73 - 9151740 CAGUUAUCGGUCAAUUU-GGUGGCUGGCCACAAAAGAGCC----------GAAAAUAAUGUGGCGCGGCUUGGUGUGGCAUUGU------------------- ((((((((((.....))-))))))))((((((...(((((----------(..............))))))..)))))).....------------------- ( -30.04, z-score = -3.71, R) >droPer1.super_11 2627098 72 - 2846995 CAGUUAUCGGUCAAUUU-GGUGGCUGGCCACAAA-GAGCC----------GAAAAUAAUGUGGCGCGGCUUGGUGUGGCAUUGU------------------- ((((((((((.....))-))))))))((((((..-(((((----------(..............))))))..)))))).....------------------- ( -31.04, z-score = -4.07, R) >droWil1.scaffold_181150 1949560 90 + 4952429 CAGUUAGCGGUCAAUUUUGGUGGCUGGCCACAAAAGAGCCCC--------GAAAAUAAUGUGGUGUC-CAUGGUGUGGCAUUGUGCC-CA-GUUGCCCACC-- ((((((.(((......))).))))))((((((..........--------........))))))...-...((((.(((((((....-))-).))))))))-- ( -25.67, z-score = 0.51, R) >droVir3.scaffold_12928 5475323 102 + 7717345 CAGUUAGCGGUCAAUUUUGAUGGCCUGCCACAAAUGAGACCCCCUUUUCUAAAUAUAAUGCGGUGUU-UUUGGUGUGGCAUUGUGCAGCACACCAACUGCAGC ......((((((...((((.(((....)))))))...)))).................((((((...-..(((((((((.....))..))))))))))))))) ( -30.00, z-score = -0.76, R) >consensus CAGUUAACGGCCAAUUU_GUUGGCCGGCCGCAAAAGAGCC__________GAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUC_AG_GCCUUCCUC___ ........(((((.......))))).((((((..........................))))))(((((.....)))))........................ (-20.00 = -19.12 + -0.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:48 2011