
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,366,792 – 4,366,899 |
| Length | 107 |
| Max. P | 0.935354 |

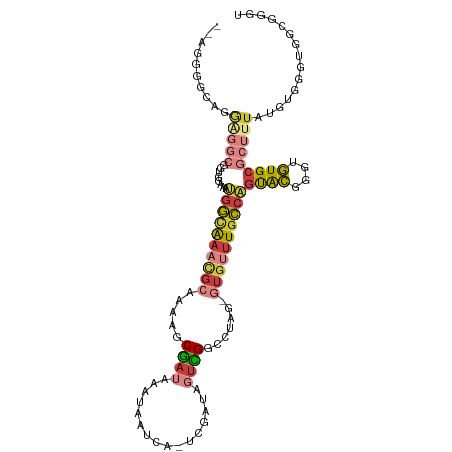
| Location | 4,366,792 – 4,366,899 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 66.36 |
| Shannon entropy | 0.69332 |
| G+C content | 0.50876 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -12.81 |
| Energy contribution | -14.29 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
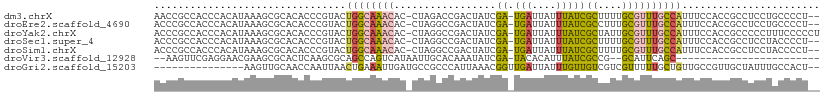
>dm3.chrX 4366792 107 + 22422827 --AGGGGCAGGAGGCGGUGGAAAUGGCAAACGCAAAAGCGAUAAAUAAUCA-UCGAUAGUCGGUCUAG-GUGUUUGCCAGUACGGGUGUGCGCUUUAUGUGGGUGGCGGUU --....(((.(((((........((((((((((......(((.....)))(-(((.....))))....-))))))))))((((....))))))))).)))........... ( -30.00, z-score = -0.84, R) >droEre2.scaffold_4690 1742779 107 + 18748788 --AGGGGCAGGAGGCGGUGGAAAUGGCAAACGCAAAGGCGAUAAAUAAUCA-UCGAUAGUCGGCCUAG-GUGUUUGCCAGUACGGGUGUGCGCUUUAUGUGGGUGGCGGGU --....(((.(((((........((((((((((..(((((((.....))).-.((.....))))))..-))))))))))((((....))))))))).)))........... ( -36.40, z-score = -2.24, R) >droYak2.chrX 17699551 109 - 21770863 AGGGGGAAAGGGGGCGGUGGAAAUGGCAAACGCAAUAGCGAUAAAUAAUCA-UCGAUAGUCGGCCUAG-GUGUUUGCCAGUACGGGUGUGCGCUUUAUGUGGGUGGCGGGU ......((((.(.(((.(.(...((((((((((....(((((.....))).-.((.....))))....-))))))))))...).).))).).))))............... ( -29.70, z-score = -0.26, R) >droSec1.super_4 3819611 107 - 6179234 --AGGGGUAGGAGGCGGUGGAAAUGGCAAACGCAAAAGCGAUAAAUAAUCA-UCGAUAGUCGGCCUAG-GUGUUUGCCAGUACGGGUGUGCGCUUUAUGUGGGUGGCGGGU --........(((((........((((((((((....(((((.....))).-.((.....))))....-))))))))))((((....)))))))))............... ( -27.90, z-score = 0.34, R) >droSim1.chrX 3373396 107 + 17042790 --AGGGGUAGGAGGCGGUGGAAAUGGCAAACGCAAAAGCGAUAAAUAAUCA-UCGAUAGUCGGCCUAG-GUGUUUGCCAGUACGGGUGUGCGCUUUAUGUGGGUGGCGGGU --........(((((........((((((((((....(((((.....))).-.((.....))))....-))))))))))((((....)))))))))............... ( -27.90, z-score = 0.34, R) >droVir3.scaffold_12928 5423769 82 + 7717345 ------------------------GCUGAAUGC--CGGCGAUAAAUGUGUA-UCGAUAUUUGUGCAAUUAUGACUGGCUGCGCUUGAGUGCGCUUCGUUCCUCGAACUU-- ------------------------...(((.((--(((..((((.((..((-........))..)).))))..))))).((((......)))))))((((...))))..-- ( -22.80, z-score = -1.12, R) >droGri2.scaffold_15203 8659888 94 - 11997470 --AGUGGCAAAUAGCAACGGCAACAGCAAAAACGACGACAACAAAUAAUCAACCGUUUAAUGGGCGGCAUCAAUUUCAGUUAAUUGGUUGCAACUU--------------- --.((.((.....)).)).(((((............................(((((.....)))))...(((((......)))))))))).....--------------- ( -16.10, z-score = 0.16, R) >consensus __AGGGGCAGGAGGCGGUGGAAAUGGCAAACGCAAAAGCGAUAAAUAAUCA_UCGAUAGUCGGCCUAG_GUGUUUGCCAGUACGGGUGUGCGCUUUAUGUGGGUGGCGGGU ..........(((((........((((((((((......(((.....)))..((((...))))......))))))))))((((....)))))))))............... (-12.81 = -14.29 + 1.48)

| Location | 4,366,792 – 4,366,899 |
|---|---|
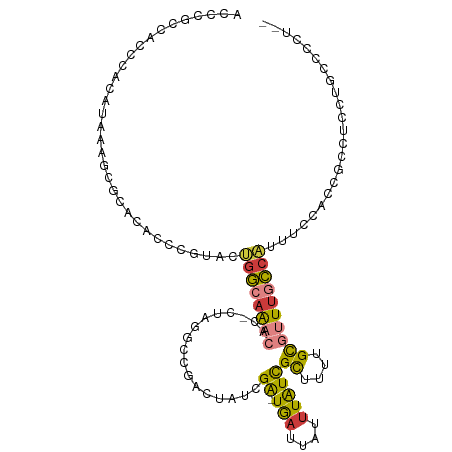
| Length | 107 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 66.36 |
| Shannon entropy | 0.69332 |
| G+C content | 0.50876 |
| Mean single sequence MFE | -19.81 |
| Consensus MFE | -6.86 |
| Energy contribution | -7.56 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4366792 107 - 22422827 AACCGCCACCCACAUAAAGCGCACACCCGUACUGGCAAACAC-CUAGACCGACUAUCGA-UGAUUAUUUAUCGCUUUUGCGUUUGCCAUUUCCACCGCCUCCUGCCCCU-- ....((............(((......)))..((((((((..-..............((-(((....)))))((....))))))))))........))...........-- ( -19.30, z-score = -2.40, R) >droEre2.scaffold_4690 1742779 107 - 18748788 ACCCGCCACCCACAUAAAGCGCACACCCGUACUGGCAAACAC-CUAGGCCGACUAUCGA-UGAUUAUUUAUCGCCUUUGCGUUUGCCAUUUCCACCGCCUCCUGCCCCU-- ....((............(((......)))..((((((((.(-..((((.(.....)((-(((....)))))))))..).))))))))........))...........-- ( -23.10, z-score = -2.68, R) >droYak2.chrX 17699551 109 + 21770863 ACCCGCCACCCACAUAAAGCGCACACCCGUACUGGCAAACAC-CUAGGCCGACUAUCGA-UGAUUAUUUAUCGCUAUUGCGUUUGCCAUUUCCACCGCCCCCUUUCCCCCU ....((............(((......)))..((((((((..-.(((.....)))..((-(((....)))))((....))))))))))........))............. ( -19.30, z-score = -1.86, R) >droSec1.super_4 3819611 107 + 6179234 ACCCGCCACCCACAUAAAGCGCACACCCGUACUGGCAAACAC-CUAGGCCGACUAUCGA-UGAUUAUUUAUCGCUUUUGCGUUUGCCAUUUCCACCGCCUCCUACCCCU-- ....((............(((......)))..((((((((.(-..((((.(.....)((-(((....)))))))))..).))))))))........))...........-- ( -20.40, z-score = -2.24, R) >droSim1.chrX 3373396 107 - 17042790 ACCCGCCACCCACAUAAAGCGCACACCCGUACUGGCAAACAC-CUAGGCCGACUAUCGA-UGAUUAUUUAUCGCUUUUGCGUUUGCCAUUUCCACCGCCUCCUACCCCU-- ....((............(((......)))..((((((((.(-..((((.(.....)((-(((....)))))))))..).))))))))........))...........-- ( -20.40, z-score = -2.24, R) >droVir3.scaffold_12928 5423769 82 - 7717345 --AAGUUCGAGGAACGAAGCGCACUCAAGCGCAGCCAGUCAUAAUUGCACAAAUAUCGA-UACACAUUUAUCGCCG--GCAUUCAGC------------------------ --..((((...))))(((((((......)))).(((.((((....)).))......(((-((......)))))..)--)).)))...------------------------ ( -19.00, z-score = -1.64, R) >droGri2.scaffold_15203 8659888 94 + 11997470 ---------------AAGUUGCAACCAAUUAACUGAAAUUGAUGCCGCCCAUUAAACGGUUGAUUAUUUGUUGUCGUCGUUUUUGCUGUUGCCGUUGCUAUUUGCCACU-- ---------------((((.(((((.(((((((((...((((((.....)))))).)))))))))...................((....)).))))).))))......-- ( -17.20, z-score = 0.07, R) >consensus ACCCGCCACCCACAUAAAGCGCACACCCGUACUGGCAAACAC_CUAGGCCGACUAUCGA_UGAUUAUUUAUCGCUUUUGCGUUUGCCAUUUCCACCGCCUCCUGCCCCU__ ................................((((((((.....................(((.....)))((....))))))))))....................... ( -6.86 = -7.56 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:47 2011