
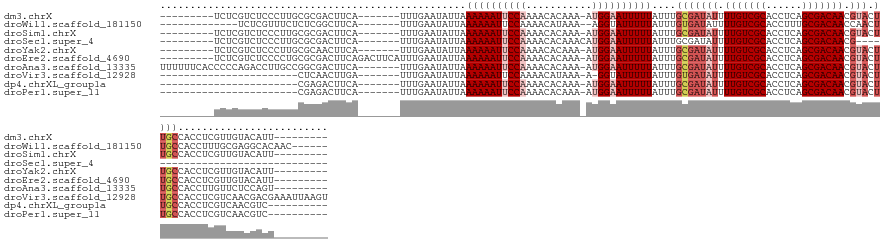
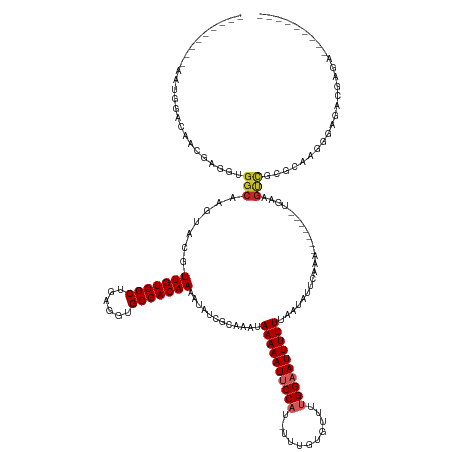
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,349,708 – 4,349,830 |
| Length | 122 |
| Max. P | 0.999491 |

| Location | 4,349,708 – 4,349,830 |
|---|---|
| Length | 122 |
| Sequences | 10 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 77.67 |
| Shannon entropy | 0.43171 |
| G+C content | 0.37704 |
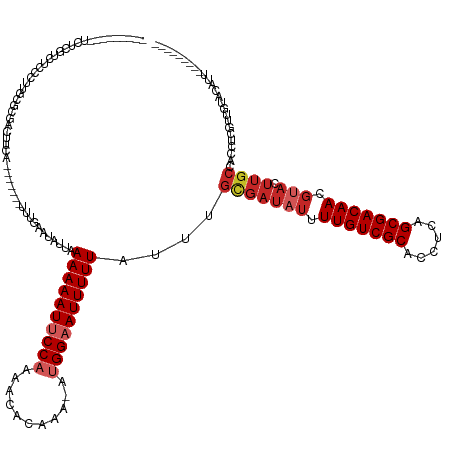
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -18.22 |
| Energy contribution | -19.28 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4349708 122 + 22422827 ---------UCUCGUCUCCCUUGCGCGACUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACACAAA-AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUUGUACAUU--------- ---------............((..((((....-------...........((((((((((.........-.))))))))))....(((((((.(((((((......))))))).))).))))......))))..))..--------- ( -28.50, z-score = -3.87, R) >droWil1.scaffold_181150 1992723 120 + 4952429 -------------UCUCGUUUCUCUCGGCUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACAUAAA--AGGUAUUUUUAUUUGUGAUAUUUUGUCGCACCUUUGCGACAACCAACUUGCCACCUUUGCGAGGCACAAC------ -------------((((((.......(((....-------...((((((((((((((.((..........--.)).)))))).....))))))))(((((((....)))))))........)))......))))))......------ ( -25.22, z-score = -2.42, R) >droSim1.chrX 3356579 122 + 17042790 ---------UCUCGUCUCCCUUGCGCGACUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACACAAA-AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUUGUACAUU--------- ---------............((..((((....-------...........((((((((((.........-.))))))))))....(((((((.(((((((......))))))).))).))))......))))..))..--------- ( -28.50, z-score = -3.87, R) >droSec1.super_4 3802539 100 - 6179234 ---------UCUCGUCUCCCUUGCGCGACUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACACAAACAUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACG-------------------------------- ---------..............(((((.(((.-------...))).....((((((((((...........))))))))))..))))).....(((((((......)))))))..-------------------------------- ( -22.40, z-score = -3.54, R) >droYak2.chrX 17682556 122 - 21770863 ---------UCUCGUCUCCCUUGCGCAACUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACACAAA-AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUUGUACAUU--------- ---------............((..((((....-------...........((((((((((.........-.))))))))))....(((((((.(((((((......))))))).))).))))......))))..))..--------- ( -28.80, z-score = -4.43, R) >droEre2.scaffold_4690 1725749 129 + 18748788 ---------UCUCGUCUCCCCUGCGCGACUUCAGACUUCAUUUGAAUAUUAAAAAAUUCCAAAACACAAA-AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUUGUACAUU--------- ---------............((..((((((((((.....)))))).....((((((((((.........-.))))))))))....(((((((.(((((((......))))))).))).))))......))))..))..--------- ( -30.00, z-score = -3.87, R) >droAna3.scaffold_13335 104271 131 + 3335858 UUUUUUCACCCCCAGACCUUGCCGGCGACUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACACAAA-AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUUGUUCUCCAGU--------- ..............((..(((....)))..)).-------...(((((...((((((((((.........-.))))))))))....(((((((.(((((((......))))))).))).)))).....)))))......--------- ( -26.60, z-score = -2.73, R) >droVir3.scaffold_12928 2326738 116 - 7717345 -----------------------CUCAACUUGA-------UUUGAAUAUUAAAAAAUUCCAAAACAUAAA-A-GGUAUUUUUAUUUGUGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUCAACGACGAAAUUAAGU -----------------------....((((((-------(((........((((((.((..........-.-)).))))))....(..((((.(((((((......))))))).))).)..).....((((...))))))))))))) ( -23.10, z-score = -2.36, R) >dp4.chrXL_group1a 3697824 107 + 9151740 -----------------------CGAGACUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACACAAA-AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUCAACGUC---------- -----------------------((((......-------...........((((((((((.........-.))))))))))....(((((((.(((((((......))))))).))).))))...))))........---------- ( -26.70, z-score = -4.09, R) >droPer1.super_11 258369 107 + 2846995 -----------------------CGAGACUUCA-------UUUGAAUAUUAAAAAAUUCCAAAACACAAA-AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUCAACGUC---------- -----------------------((((......-------...........((((((((((.........-.))))))))))....(((((((.(((((((......))))))).))).))))...))))........---------- ( -26.70, z-score = -4.09, R) >consensus _________UCUCGUCUCCCUUGCGCGACUUCA_______UUUGAAUAUUAAAAAAUUCCAAAACACAAA_AUGGAAUUUUUAUUUGCGAUAUUUUGUCGCACCUCAGCGACAACGUACUUGCCACCUCGUUGUACAUU_________ ...................................................((((((((((...........))))))))))....(((((((.(((((((......))))))).))).))))......................... (-18.22 = -19.28 + 1.06)
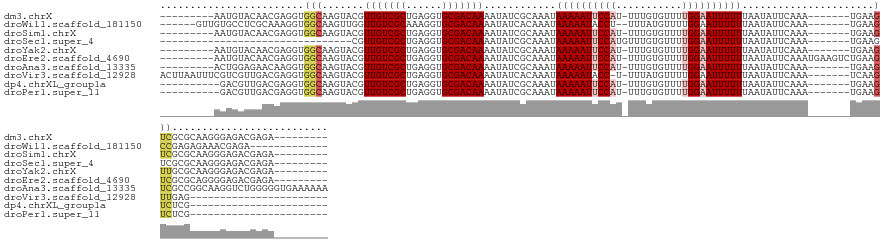
| Location | 4,349,708 – 4,349,830 |
|---|---|
| Length | 122 |
| Sequences | 10 |
| Columns | 148 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Shannon entropy | 0.43171 |
| G+C content | 0.37704 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4349708 122 - 22422827 ---------AAUGUACAACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU-UUUGUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGUCGCGCAAGGGAGACGAGA--------- ---------.........(..((((((((.....))))))))..)(((((((..............((((((((((.-.........)))))))))).....(((...-------.))))))))))...(....)....--------- ( -32.10, z-score = -1.86, R) >droWil1.scaffold_181150 1992723 120 - 4952429 ------GUUGUGCCUCGCAAAGGUGGCAAGUUGGUUGUCGCAAAGGUGCGACAAAAUAUCACAAAUAAAAAUACCU--UUUAUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGCCGAGAGAAACGAGA------------- ------.((((..(((.((((((((.....(((.((((((((....)))))))).......))).......)))))--))...(((((.((((........))))...-------.))))).).)))..))))..------------- ( -26.20, z-score = -0.63, R) >droSim1.chrX 3356579 122 - 17042790 ---------AAUGUACAACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU-UUUGUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGUCGCGCAAGGGAGACGAGA--------- ---------.........(..((((((((.....))))))))..)(((((((..............((((((((((.-.........)))))))))).....(((...-------.))))))))))...(....)....--------- ( -32.10, z-score = -1.86, R) >droSec1.super_4 3802539 100 + 6179234 --------------------------------CGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAUGUUUGUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGUCGCGCAAGGGAGACGAGA--------- --------------------------------((((.((.((...(((((((..............((((((((((...........)))))))))).....(((...-------.)))))))))).)).))))))...--------- ( -28.20, z-score = -2.40, R) >droYak2.chrX 17682556 122 + 21770863 ---------AAUGUACAACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU-UUUGUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGUUGCGCAAGGGAGACGAGA--------- ---------..((..((((..((((((((.....))))))))(((.(((((.......)))))...((((((((((.-.........)))))))))).....)))...-------....))))..))..(....)....--------- ( -30.20, z-score = -1.38, R) >droEre2.scaffold_4690 1725749 129 - 18748788 ---------AAUGUACAACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU-UUUGUGUUUUGGAAUUUUUUAAUAUUCAAAUGAAGUCUGAAGUCGCGCAGGGGAGACGAGA--------- ---------.........(..((((((((.....))))))))..)(((((((..............((((((((((.-.........)))))))))).....(((....))).......)))))))...(....)....--------- ( -32.10, z-score = -1.02, R) >droAna3.scaffold_13335 104271 131 - 3335858 ---------ACUGGAGAACAAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU-UUUGUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGUCGCCGGCAAGGUCUGGGGGUGAAAAAA ---------.(..((......((((((..(((..(((((((......)))))))..))).......((((((((((.-.........))))))))))...........-------....)))))).......))..)........... ( -33.12, z-score = -1.30, R) >droVir3.scaffold_12928 2326738 116 + 7717345 ACUUAAUUUCGUCGUUGACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCACAAAUAAAAAUACC-U-UUUAUGUUUUGGAAUUUUUUAAUAUUCAAA-------UCAAGUUGAG----------------------- .((((((((.(((((...(..((((((((.....))))))))..)..)))))(((((.(((...((((((.....-)-)))))....))).)))))............-------..))))))))----------------------- ( -24.50, z-score = -1.02, R) >dp4.chrXL_group1a 3697824 107 - 9151740 ----------GACGUUGACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU-UUUGUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGUCUCG----------------------- ----------(((.(((((..((((((((.....))))))))..).(((((.......)))))...((((((((((.-.........))))))))))......)))).-------....)))...----------------------- ( -26.10, z-score = -1.30, R) >droPer1.super_11 258369 107 - 2846995 ----------GACGUUGACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU-UUUGUGUUUUGGAAUUUUUUAAUAUUCAAA-------UGAAGUCUCG----------------------- ----------(((.(((((..((((((((.....))))))))..).(((((.......)))))...((((((((((.-.........))))))))))......)))).-------....)))...----------------------- ( -26.10, z-score = -1.30, R) >consensus _________AAUGGACAACGAGGUGGCAAGUACGUUGUCGCUGAGGUGCGACAAAAUAUCGCAAAUAAAAAUUCCAU_UUUGUGUUUUGGAAUUUUUUAAUAUUCAAA_______UGAAGUCGCGCAAGGGAGACGAGA_________ ........................(((.......(((((((......)))))))............((((((((((...........))))))))))......................))).......................... (-18.25 = -18.73 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:45 2011