| Sequence ID | dm3.chr2L |
|---|---|
| Location | 889,491 – 889,592 |
| Length | 101 |
| Max. P | 0.958487 |

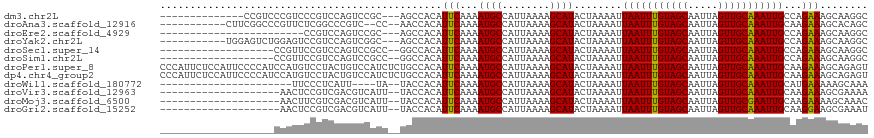
| Location | 889,491 – 889,592 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Shannon entropy | 0.45396 |
| G+C content | 0.37727 |
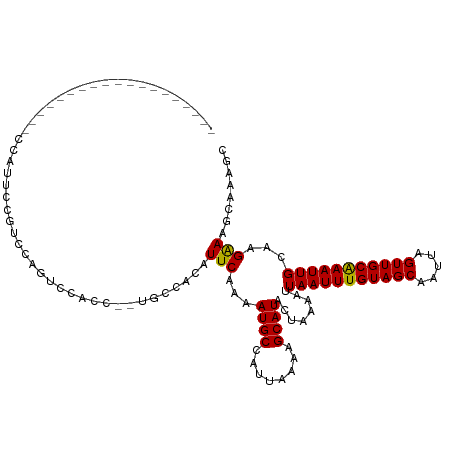
| Mean single sequence MFE | -18.17 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.22 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958487 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 889491 101 + 23011544 --------------CCGUCCCGUCCCGUCCAGUCCGC---AGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCCAGAAAGCAAGGC --------------.......................---.(((.........((((........))))........(((((((((((.....)))))))))))...........))) ( -16.30, z-score = -1.03, R) >droAna3.scaffold_12916 2612602 103 + 16180835 -----------CUUCGGCCCGUUCUCGGCCCGUC--CC--AACCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCAAGAAAGCACAGC -----------...(((.(((....))).)))..--..--.......(((...((((........))))........(((((((((((.....)))))))))))...)))........ ( -20.90, z-score = -1.76, R) >droEre2.scaffold_4929 947671 91 + 26641161 ------------------------CCGUCCAGUCCGC---AGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCCAGAAAGCAAGGC ------------------------.............---.(((.........((((........))))........(((((((((((.....)))))))))))...........))) ( -16.30, z-score = -1.14, R) >droYak2.chr2L 874158 104 + 22324452 -----------UGGAGUCUGGAGUCCGUCCAGUCGGC---AGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCCAGAAAGCAAGGC -----------....(.(((((.....))))).)(((---.............((((........)))).........((((((((((.....)))))))))))))............ ( -24.20, z-score = -1.21, R) >droSec1.super_14 860139 97 + 2068291 -------------------CCGUUCCGUCCAGUCCGCC--GGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCCAGAAAGCAAGGC -------------------................(((--(((..........((((........)))).........((((((((((.....))))))))))))).(....)..))) ( -18.10, z-score = -0.83, R) >droSim1.chr2L 875460 97 + 22036055 -------------------CCGUUCCGUCCAGUCCGCC--GGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCCAGAAAGCAAGGC -------------------................(((--(((..........((((........)))).........((((((((((.....))))))))))))).(....)..))) ( -18.10, z-score = -0.83, R) >droPer1.super_8 3440462 118 + 3966273 CCCAUUCUCCAUUCCCCAUCCAUGUCCUACUGUCCAUCUCUGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCAAGAAAGCAGAGU .....................(((..(....)..)))((((((....(((...((((........))))........(((((((((((.....)))))))))))...))).)))))). ( -22.00, z-score = -3.51, R) >dp4.chr4_group2 786585 118 - 1235136 CCCAUUCUCCAUUCCCCAUCCAUGUCCUACUGUCCAUCUCUGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCAAGAAAGCAGAGU .....................(((..(....)..)))((((((....(((...((((........))))........(((((((((((.....)))))))))))...))).)))))). ( -22.00, z-score = -3.51, R) >droWil1.scaffold_180772 6668728 90 - 8906247 ----------------------UUCCCUCAUU----UA--UACCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCAUGAAAAAGCAAA ----------------------..........----..--.......((((..((((........))))........(((((((((((.....)))))))))))..))))........ ( -14.10, z-score = -1.37, R) >droVir3.scaffold_12963 19444577 96 + 20206255 --------------------AACUCCGUCGACGUCAUU--UACCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCAAGAAAGCGAAAA --------------------.......(((...((...--.............((((........))))........(((((((((((.....)))))))))))...))...)))... ( -16.60, z-score = -1.86, R) >droMoj3.scaffold_6500 8580110 96 - 32352404 --------------------AACUUCGUCGACGUCAUU--UACCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCGAAUUGCAAGAAAAGCAAAC --------------------...((((.(((((.((((--(..........))))).).......((.(((............))))).....))))))))((((.......)))).. ( -13.30, z-score = -0.38, R) >droGri2.scaffold_15252 7304045 96 + 17193109 --------------------AACUCCGUCGACGUCAUU--UACCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCAAGGAAGCGAAAU --------------------.......(((...((...--.............((((........))))........(((((((((((.....)))))))))))....))..)))... ( -16.10, z-score = -1.21, R) >consensus ___________________CCAUUCCGUCCAGUCCACC__UGCCACAUUCAAAAUGCCAUUAAAAGCAUACUAAAAUUAAUUUGUAGCAAUUAGUUGCAAAUUGCAAGAAAGCAAAGC ...............................................(((...((((........))))........(((((((((((.....)))))))))))...)))........ (-13.37 = -13.22 + -0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:16 2011