
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,298,430 – 4,298,495 |
| Length | 65 |
| Max. P | 0.997000 |

| Location | 4,298,430 – 4,298,495 |
|---|---|
| Length | 65 |
| Sequences | 7 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 89.49 |
| Shannon entropy | 0.19516 |
| G+C content | 0.47555 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.997000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
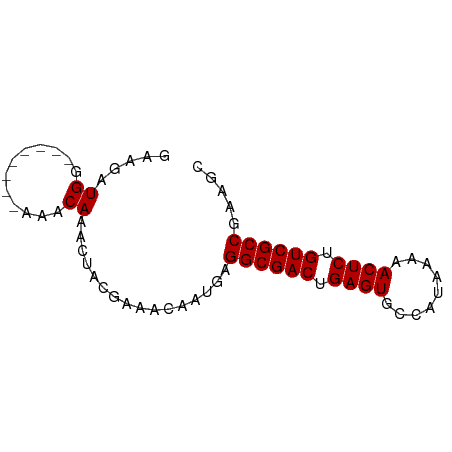
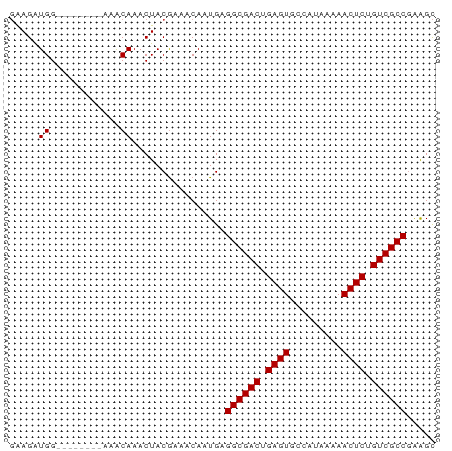
>dm3.chrX 4298430 65 + 22422827 GAAGAUGG--------AAGCAAACUACGAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCGAAGC ........--------.....................((((((.((((.........)))).))))))..... ( -16.90, z-score = -2.80, R) >droSec1.super_4 3752235 65 - 6179234 GAAGAUGG--------AAACAAACUACGAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCGAAGC .....((.--------...))................((((((.((((.........)))).))))))..... ( -19.40, z-score = -4.56, R) >droYak2.chrX 17630353 65 - 21770863 GAAGAUGG--------AAACAAACUACGAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCGAAGC .....((.--------...))................((((((.((((.........)))).))))))..... ( -19.40, z-score = -4.56, R) >droEre2.scaffold_4690 1673496 65 + 18748788 GAAGAUGG--------AAGCAAACUACGAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCGAGGC ........--------.....................((((((.((((.........)))).))))))..... ( -16.90, z-score = -2.33, R) >droAna3.scaffold_13335 3169402 67 - 3335858 -GAGGUGGUGG-----CAACAAACUACAAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCAGAGU -...((((((.-----...)..)))))..........((((((.((((.........)))).))))))..... ( -22.10, z-score = -3.15, R) >dp4.chrXL_group1a 5390940 73 - 9151740 GAAGAUGGCAGUGGAGAAACAAACUACGAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCGAAGC .......((.((((.........))))..........((((((.((((.........)))).))))))...)) ( -19.50, z-score = -2.69, R) >droPer1.super_11 2525548 73 - 2846995 GAAGAUGGCAGUGGAGAAACAAACUACGAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCGAAGC .......((.((((.........))))..........((((((.((((.........)))).))))))...)) ( -19.50, z-score = -2.69, R) >consensus GAAGAUGG________AAACAAACUACGAAACAAUGAGGCGACUGAGUGCCAUAAAAACUCUGUCGCCGAAGC .....................................((((((.((((.........)))).))))))..... (-16.90 = -16.90 + 0.00)

| Location | 4,298,430 – 4,298,495 |
|---|---|
| Length | 65 |
| Sequences | 7 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 89.49 |
| Shannon entropy | 0.19516 |
| G+C content | 0.47555 |
| Mean single sequence MFE | -15.61 |
| Consensus MFE | -14.46 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
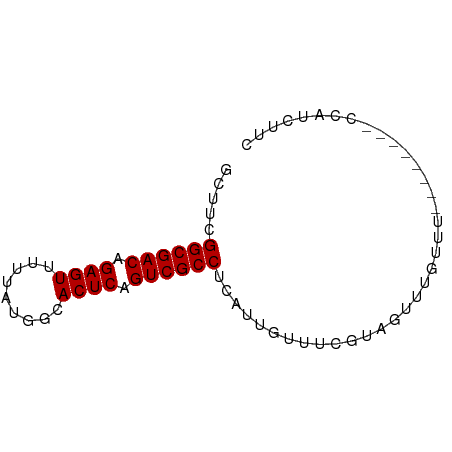
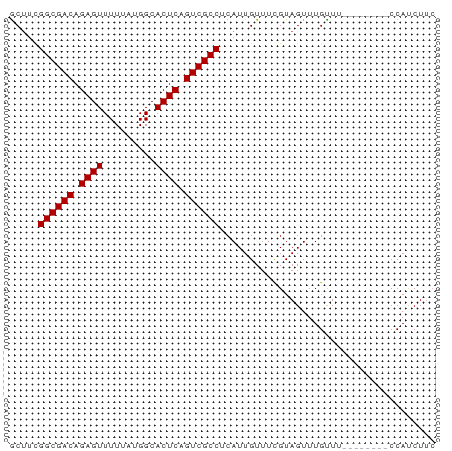
>dm3.chrX 4298430 65 - 22422827 GCUUCGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUCGUAGUUUGCUU--------CCAUCUUC .....((((((.((((.........)))).)))))).....................--------........ ( -14.50, z-score = -1.49, R) >droSec1.super_4 3752235 65 + 6179234 GCUUCGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUCGUAGUUUGUUU--------CCAUCUUC .....((((((.((((.........)))).)))))).....................--------........ ( -14.50, z-score = -1.91, R) >droYak2.chrX 17630353 65 + 21770863 GCUUCGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUCGUAGUUUGUUU--------CCAUCUUC .....((((((.((((.........)))).)))))).....................--------........ ( -14.50, z-score = -1.91, R) >droEre2.scaffold_4690 1673496 65 - 18748788 GCCUCGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUCGUAGUUUGCUU--------CCAUCUUC .....((((((.((((.........)))).)))))).....................--------........ ( -14.50, z-score = -1.40, R) >droAna3.scaffold_13335 3169402 67 + 3335858 ACUCUGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUUGUAGUUUGUUG-----CCACCACCUC- ((((((....)))))).....(((((..(((..((..((......))..))))).))-----))).......- ( -17.30, z-score = -1.75, R) >dp4.chrXL_group1a 5390940 73 + 9151740 GCUUCGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUCGUAGUUUGUUUCUCCACUGCCAUCUUC .....((((((.((((.........)))).))))))..........(((((..........)))))....... ( -17.00, z-score = -1.93, R) >droPer1.super_11 2525548 73 + 2846995 GCUUCGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUCGUAGUUUGUUUCUCCACUGCCAUCUUC .....((((((.((((.........)))).))))))..........(((((..........)))))....... ( -17.00, z-score = -1.93, R) >consensus GCUUCGGCGACAGAGUUUUUAUGGCACUCAGUCGCCUCAUUGUUUCGUAGUUUGUUU________CCAUCUUC .....((((((.((((.........)))).))))))..................................... (-14.46 = -14.46 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:35 2011