| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,289,826 – 4,289,931 |
| Length | 105 |
| Max. P | 0.975895 |

| Location | 4,289,826 – 4,289,926 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.73 |
| Shannon entropy | 0.36865 |
| G+C content | 0.41092 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.27 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
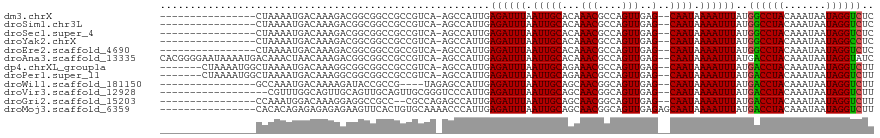
>dm3.chrX 4289826 100 + 22422827 ----------------CUAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUC ----------------.......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).. ( -30.40, z-score = -3.96, R) >droSim1.chr3L 4518628 100 + 22553184 ----------------CUAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUC ----------------.......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).. ( -30.40, z-score = -3.96, R) >droSec1.super_4 3743269 100 - 6179234 ----------------CUAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUC ----------------.......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).. ( -30.40, z-score = -3.96, R) >droYak2.chrX 17621465 100 - 21770863 ----------------CUAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGCCUC ----------------.............((((((....)))))).-........((((((.(((((...(((....)))..)--)))).))))))..((((((.......)))))).. ( -29.80, z-score = -3.62, R) >droEre2.scaffold_4690 1665127 100 + 18748788 ----------------CUAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUC ----------------.......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).. ( -30.40, z-score = -3.96, R) >droAna3.scaffold_13335 3157868 116 - 3335858 CACGGGGAAUAAAAUGACAAACUAACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUAUC .............................((((((....)))))).-...(((..((((((.(((((...(((....)))..)--)))).)))))))))(((((.......)))))... ( -24.90, z-score = -1.70, R) >dp4.chrXL_group1a 5379225 109 - 9151740 -------CUAAAAUGGCUAAAAUGACAAAGGCGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCAGAAACGCCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUU -------....(((((((....((((...(((((...)))))))))-))))))).((((((.(((((...(((....)))..)--)))).))))))..((((((.......)))))).. ( -34.70, z-score = -4.18, R) >droPer1.super_11 2512269 109 - 2846995 -------CUAAAAUGGCUAAAAUGACAAAGGCGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCAGAAACGCCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUU -------....(((((((....((((...(((((...)))))))))-))))))).((((((.(((((...(((....)))..)--)))).))))))..((((((.......)))))).. ( -34.70, z-score = -4.18, R) >droWil1.scaffold_181150 2884416 97 - 4952429 ----------------GCCAAAUGACAAAAGAUACCGCCG----UAGAGCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUU ----------------(((((.........(....)((((----(.(.((.(((((...))))).))..).)))))..))).)--)............((((((.......)))))).. ( -18.50, z-score = -1.36, R) >droVir3.scaffold_12928 5306608 99 + 7717345 ------------------CGUUUGGCAGUUGCAGUUGCAGUUGCGGGUCCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUU ------------------.(((..((.((((..((((((((((..((((.......))))))))))))))..)))).))..))--)............((((((.......)))))).. ( -29.60, z-score = -2.26, R) >droGri2.scaffold_15203 5768009 99 - 11997470 ----------------CCAAAUGGACAAAGGAGGCCGCC--CGCCAGAGCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUU ----------------...(((((.....((.((....)--).))....))))).((((((.(((((..((((....)))).)--)))).))))))..((((((.......)))))).. ( -25.50, z-score = -2.05, R) >droMoj3.scaffold_6359 3840747 103 + 4525533 ----------------CACACAGAGAGAGAGAAGUUCACUGUGCAAAACCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAGAGCAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUU ----------------..(((((.(((.......))).)))))............((((((.(((((..((((....))))...))))).))))))..((((((.......)))))).. ( -24.90, z-score = -3.18, R) >consensus ________________CUAAAAUGACAAAGACGGCGGCCGCCGUCA_AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG__CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUC ...................................................((((...(((((((((......).))))))))..)))).........((((((.......)))))).. (-12.50 = -12.27 + -0.24)
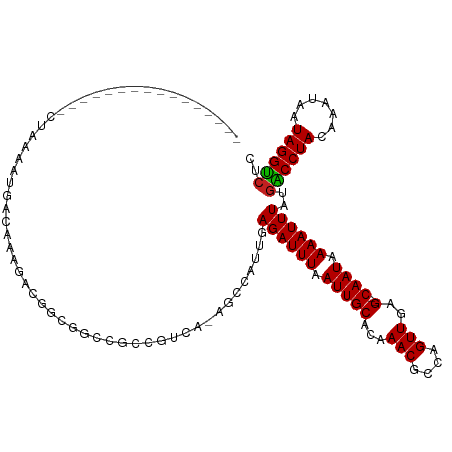
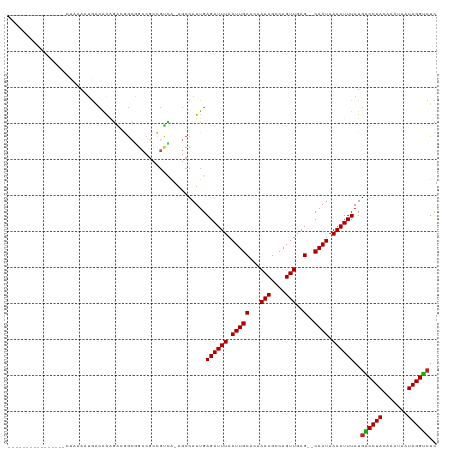
| Location | 4,289,826 – 4,289,926 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Shannon entropy | 0.36865 |
| G+C content | 0.41092 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -8.38 |
| Energy contribution | -8.18 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4289826 100 - 22422827 GAGACCUAUUAUUUGUAGGCCAUAAAUUUUAUUG--CUCAACUGGCGUUUGUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGUCUUUGUCAUUUUAG---------------- (((((((((.....))))).......(((.((((--(.(((.......))).))))).)))))))((((((.-.((((((....))))))...))))))....---------------- ( -31.20, z-score = -3.27, R) >droSim1.chr3L 4518628 100 - 22553184 GAGACCUAUUAUUUGUAGGCCAUAAAUUUUAUUG--CUCAACUGGCGUUUGUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGUCUUUGUCAUUUUAG---------------- (((((((((.....))))).......(((.((((--(.(((.......))).))))).)))))))((((((.-.((((((....))))))...))))))....---------------- ( -31.20, z-score = -3.27, R) >droSec1.super_4 3743269 100 + 6179234 GAGACCUAUUAUUUGUAGGCCAUAAAUUUUAUUG--CUCAACUGGCGUUUGUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGUCUUUGUCAUUUUAG---------------- (((((((((.....))))).......(((.((((--(.(((.......))).))))).)))))))((((((.-.((((((....))))))...))))))....---------------- ( -31.20, z-score = -3.27, R) >droYak2.chrX 17621465 100 + 21770863 GAGGCCUAUUAUUUGUAGGCCAUAAAUUUUAUUG--CUCAACUGGCGUUUGUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGUCUUUGUCAUUUUAG---------------- ..(((((((.....)))))))....((((.((((--(.(((.......))).))))).))))...((((((.-.((((((....))))))...))))))....---------------- ( -35.20, z-score = -4.07, R) >droEre2.scaffold_4690 1665127 100 - 18748788 GAGACCUAUUAUUUGUAGGCCAUAAAUUUUAUUG--CUCAACUGGCGUUUGUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGUCUUUGUCAUUUUAG---------------- (((((((((.....))))).......(((.((((--(.(((.......))).))))).)))))))((((((.-.((((((....))))))...))))))....---------------- ( -31.20, z-score = -3.27, R) >droAna3.scaffold_13335 3157868 116 + 3335858 GAUACCUAUUAUUUGUAGGUCAUAAAUUUUAUUG--CUCAACUGGCGUUUGUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGUCUUUGUUAGUUUGUCAUUUUAUUCCCCGUG (((((((((.....)))))).....((((.((((--(.(((.......))).))))).)))).....((((.-.((((((....))))))...))))....)))............... ( -26.10, z-score = -1.16, R) >dp4.chrXL_group1a 5379225 109 + 9151740 AAGACCUAUUAUUUGUAGGUCAUAAAUUUUAUUG--CUCAACUGGCGUUUCUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGCCUUUGUCAUUUUAGCCAUUUUAG------- ..(((((((.....)))))))....((((.((((--(..(((....)))...))))).))))...(((((((-(((((((((...)))))...))))....)))))))....------- ( -33.30, z-score = -3.81, R) >droPer1.super_11 2512269 109 + 2846995 AAGACCUAUUAUUUGUAGGUCAUAAAUUUUAUUG--CUCAACUGGCGUUUCUGCAAUUAAAUCUCAAUGGCU-UGACGGCGGCCGCCGCCUUUGUCAUUUUAGCCAUUUUAG------- ..(((((((.....)))))))....((((.((((--(..(((....)))...))))).))))...(((((((-(((((((((...)))))...))))....)))))))....------- ( -33.30, z-score = -3.81, R) >droWil1.scaffold_181150 2884416 97 + 4952429 AAGACCUAUUAUUUGUAGGUCAUAAAUUUUAUUG--CUCAACUGCCGUUGCUGCAAUUAAAUCUCAAUGGCUCUA----CGGCGGUAUCUUUUGUCAUUUGGC---------------- ..(((((((.....)))))))(((((...(((((--((.....(((((((..............)))))))....----.)))))))...)))))........---------------- ( -25.64, z-score = -3.02, R) >droVir3.scaffold_12928 5306608 99 - 7717345 AAGACCUAUUAUUUGUAGGUCAUAAAUUUUAUUG--CUCAACUGCCGUUGCUGCAAUUAAAUCUCAAUGGGACCCGCAACUGCAACUGCAACUGCCAAACG------------------ ..(((((((.....))))))).....(((.((((--(.((((....))))..))))).)))(((.....)))...(((..(((....)))..)))......------------------ ( -23.90, z-score = -2.31, R) >droGri2.scaffold_15203 5768009 99 + 11997470 AAGACCUAUUAUUUGUAGGUCAUAAAUUUUAUUG--CUCAACUGCCGUUGCUGCAAUUAAAUCUCAAUGGCUCUGGCG--GGCGGCCUCCUUUGUCCAUUUGG---------------- ..(((((((.....)))))))....((((.((((--(.((((....))))..))))).))))((.((((((...((.(--(....)).))...).))))).))---------------- ( -27.10, z-score = -1.88, R) >droMoj3.scaffold_6359 3840747 103 - 4525533 AAGACCUAUUAUUUGUAGGUCAUAAAUUUUAUUGCUCUCAACUGCCGUUGCUGCAAUUAAAUCUCAAUGGGUUUUGCACAGUGAACUUCUCUCUCUCUGUGUG---------------- ..(((((((.....)))))))....((((.(((((...((((....))))..))))).)))).............((((((.((.........)).)))))).---------------- ( -25.90, z-score = -3.57, R) >consensus AAGACCUAUUAUUUGUAGGUCAUAAAUUUUAUUG__CUCAACUGGCGUUUCUGCAAUUAAAUCUCAAUGGCU_UGACGGCGGCCGCCGCCUUUGUCAUUUUAG________________ ..(((........(((.((((((..((((.((((.....(((....)))....)))).))))....))))))...)))..(((....)))...)))....................... ( -8.38 = -8.18 + -0.19)
| Location | 4,289,827 – 4,289,931 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Shannon entropy | 0.37857 |
| G+C content | 0.40835 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.27 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.955078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4289827 104 + 22422827 ---------UAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUCAUCCG--- ---------......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).......--- ( -30.40, z-score = -3.65, R) >droSim1.chr3L 4518629 104 + 22553184 ---------UAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUCAUCCG--- ---------......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).......--- ( -30.40, z-score = -3.65, R) >droSec1.super_4 3743270 104 - 6179234 ---------UAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUCAUCCG--- ---------......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).......--- ( -30.40, z-score = -3.65, R) >droYak2.chrX 17621466 104 - 21770863 ---------UAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGCCUCAUCCG--- ---------....((((....((((((....)))))).-........((((((.(((((...(((....)))..)--)))).))))))..((((((.......))))))))))...--- ( -30.40, z-score = -3.57, R) >droEre2.scaffold_4690 1665128 104 + 18748788 ---------UAAAAUGACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGGCCUACAAAUAAUAGGUCUCAUCCG--- ---------......(((...((((((....)))))).-.(((((..((((((.(((((...(((....)))..)--)))).))))))))))).............))).......--- ( -30.40, z-score = -3.65, R) >droAna3.scaffold_13335 3157876 113 - 3335858 AUAAAAUGACAAACUAACAAAGACGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUAUCAUCCA--- .....((((............((((((....)))))).-...(((..((((((.(((((...(((....)))..)--)))).)))))))))(((((.......))))).))))...--- ( -25.70, z-score = -2.86, R) >dp4.chrXL_group1a 5379226 113 - 9151740 UAAAAUGGCUAAAAUGACAAAGGCGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCAGAAACGCCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUUGUUCG--- ...(((((((....((((...(((((...)))))))))-))))))).((((((.(((((...(((....)))..)--)))).))))))..((((((.......)))))).......--- ( -34.70, z-score = -3.58, R) >droPer1.super_11 2512270 113 - 2846995 UAAAAUGGCUAAAAUGACAAAGGCGGCGGCCGCCGUCA-AGCCAUUGAGAUUUAAUUGCAGAAACGCCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUUGUUCG--- ...(((((((....((((...(((((...)))))))))-))))))).((((((.(((((...(((....)))..)--)))).))))))..((((((.......)))))).......--- ( -34.70, z-score = -3.58, R) >droWil1.scaffold_181150 2884417 101 - 4952429 ---------CCAAAUGACAAAAGAUACCGCCG----UAGAGCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUUGCUUA--- ---------.......................----..((((.....((((((.(((((..((((....)))).)--)))).))))))..((((((.......))))))..)))).--- ( -21.10, z-score = -1.75, R) >droVir3.scaffold_12928 5306608 104 + 7717345 ----------CGUUUGGCAGUUGCAGUUGCAGUUGCGGGUCCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUUGUUCA--- ----------.(((..((.((((..((((((((((..((((.......))))))))))))))..)))).))..))--)............((((((.......)))))).......--- ( -29.60, z-score = -1.66, R) >droGri2.scaffold_15203 5768010 103 - 11997470 ---------CAAAUGGACAAAGGAGGCCGCC--CGCCAGAGCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAG--CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUUGUUCA--- ---------....(((((((.((.(((....--.)))....))....((((((.(((((..((((....)))).)--)))).))))))..((((((.......)))))))))))))--- ( -27.90, z-score = -2.17, R) >droMoj3.scaffold_6359 3840748 110 + 4525533 ---------ACACAGAGAGAGAGAAGUUCACUGUGCAAAACCCAUUGAGAUUUAAUUGCAGCAACGGCAGUUGAGAGCAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUUGUUUAUCA ---------.(((((.(((.......))).)))))..((((......((((((.(((((..((((....))))...))))).))))))..((((((.......))))))..)))).... ( -25.60, z-score = -2.33, R) >consensus _________UAAAAUGACAAAGACGGCGGCCGCCGUCA_AGCCAUUGAGAUUUAAUUGCACAAACGCCAGUUGAG__CAAUAAAAUUUAUGACCUACAAAUAAUAGGUCUCAUCCG___ ...........................................((((...(((((((((......).))))))))..)))).........((((((.......)))))).......... (-12.50 = -12.27 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:33 2011