
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,288,736 – 4,288,824 |
| Length | 88 |
| Max. P | 0.957944 |

| Location | 4,288,736 – 4,288,824 |
|---|---|
| Length | 88 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 73.75 |
| Shannon entropy | 0.51356 |
| G+C content | 0.61259 |
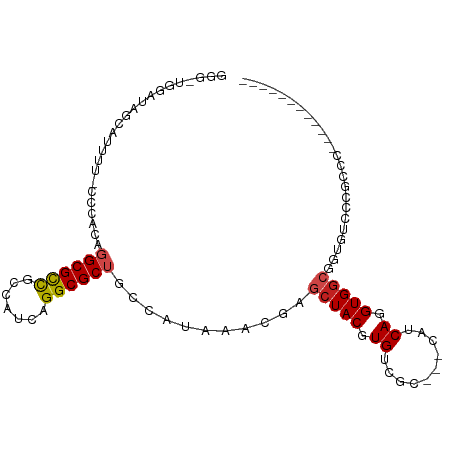
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -17.13 |
| Energy contribution | -17.39 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
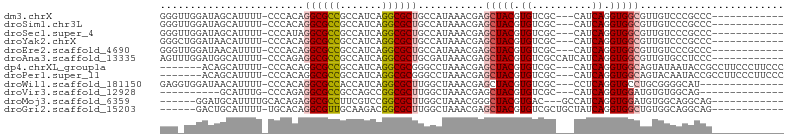
>dm3.chrX 4288736 88 - 22422827 GGGUUGGAUAGCAUUUU-CCCACAGGCGCCGCCAUCAGGCGCUGCCAUAAACGAGCUACGUGUCGC---CAUCAGGUGGCGUUGUCCCGCCC------------ ((((.(((((((.....-......((((.((((....)))).))))........(((((.((....---...)).)))))))))))).))))------------ ( -39.80, z-score = -3.12, R) >droSim1.chr3L 4517574 88 - 22553184 GGGUUGGAUAGCAUUUU-CCCACAGGCGCCGCCAUCAGGCGCUGCCAUAAACGAGCUACGUGUCGC---CAUCAGGUGGCGUUGUCCCGCCC------------ ((((.(((((((.....-......((((.((((....)))).))))........(((((.((....---...)).)))))))))))).))))------------ ( -39.80, z-score = -3.12, R) >droSec1.super_4 3742223 88 + 6179234 GGGUUGGAUAGCAUUUU-CCCAUAGGCGCCGCCAUCAGGCGCUGCCAUAAACGAGCUACGUGUCGC---CAUCAGGUGGCGUUGUCCCGCCC------------ ((((.(((((((.....-......((((.((((....)))).))))........(((((.((....---...)).)))))))))))).))))------------ ( -39.80, z-score = -3.24, R) >droYak2.chrX 17620310 88 + 21770863 GGGCUGGAUAACAUUUU-CCCACAGGCGCCGCCAUCAGGCGCUGCCAUAAACGAGCUACGUGUCGC---CAUCAGGUGGCGUUGUCCCGCCC------------ ((((.(((((((.....-......((((.((((....)))).))))........(((((.((....---...)).)))))))))))).))))------------ ( -41.40, z-score = -3.71, R) >droEre2.scaffold_4690 1664007 88 - 18748788 GGGUUGGAUAACAUUUU-CCCACAGGCGCCGCCAUCAGGCGCUGCCAUAAACGAGCUACGUGUCGC---CAUCAGGUGGCGUUGUCCCGCCC------------ ((((.(((((((.....-......((((.((((....)))).))))........(((((.((....---...)).)))))))))))).))))------------ ( -39.80, z-score = -3.45, R) >droAna3.scaffold_13335 3157033 91 + 3335858 AGUUUGGAUGGCAUUUU-CCCAGAGGCGCCGCCAUCAGGCGCUGCGAUAAACGAGCUACGUGUCGCCAUCAUCAGGUGGCGUUGUGCCUCCC------------ .....(((.(((((...-......(((((.(((....)))(((.((.....)))))...)))))((((((....))))))...)))))))).------------ ( -34.20, z-score = -1.25, R) >dp4.chrXL_group1a 5378057 93 + 9151740 -------ACAGCAUUUU-CCCACAGGCGCCGCCAUCAGGCGCGGGCCUAAACGAGCUACGUGUCGC---CAUCAGGUGGCAGUAUAAUACCGCCUUCCCUUCCC -------...((.....-.....((((.(((((....)))).).))))........(((.((((((---(....)))))))))).......))........... ( -28.50, z-score = -0.96, R) >droPer1.super_11 2511101 93 + 2846995 -------ACAGCAUUUU-CCCACAGGCGCCGCCAUCAGGCGCGGGCCUAAACGAGCUACGUGUCGC---CAUCAGGUGGCAGUACAAUACCGCCUUCCCUUCCC -------...((.....-.....((((.(((((....)))).).))))......(.(((.((((((---(....)))))))))))......))........... ( -28.60, z-score = -1.01, R) >droWil1.scaffold_181150 2883848 86 + 4952429 GAGGUGGAUAACAUUUU-CCCACAGGCGCCACCAUCAGGCGCUUGGCUAAACGAGCUACGUGUCGC---CCUCAGGUGCCUGCGGGGCAU-------------- ((((((.....))))))-(((.((((((((.......(((((.(((((.....))))).)))))..---.....)))))))).)))....-------------- ( -37.44, z-score = -2.42, R) >droVir3.scaffold_12928 5305168 76 - 7717345 ----------GCAUUUG-CCCAGAGGCGCCGCCAGCCGGCGCUUGGCUAAACGAGCUACGUGUCGC---CAUCAGGUGGAUGUGUGGCAG-------------- ----------......(-((...((((((((.....))))))))))).......((((((..((((---(....))).))..))))))..-------------- ( -32.70, z-score = -1.11, R) >droMoj3.scaffold_6359 3839511 83 - 4525533 ------GGAUGCAUUUUGCACAGAGGCGCCUUCGUCCGGCGCUUGGCUAAACGGGCUACGUGAC---GCCAUCAGGUGGAUGUGGCAGGCAG------------ ------...(((.....((......))(((..((((((.(...((((...(((.....)))...---))))...).)))))).)))..))).------------ ( -30.40, z-score = -0.34, R) >droGri2.scaffold_15203 5766956 85 + 11997470 ------GACUGCAUUUU-UGCACAGGCGUUGCAAGACGGCGCUUGGCUAAACGAGCUACGUGUCGCUGCUAUCAGGUGGCUGUGGCAGGCAG------------ ------..((((.....-.((.(((((((((.....))))))))))).......((((((.((((((.......))))))))))))..))))------------ ( -35.00, z-score = -1.57, R) >consensus GGG_UGGAUAGCAUUUU_CCCACAGGCGCCGCCAUCAGGCGCUGCCAUAAACGAGCUACGUGUCGC___CAUCAGGUGGCGGUGUCCCGCCC____________ ........................((((((.......))))))...........(((((.((..........)).)))))........................ (-17.13 = -17.39 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:30 2011