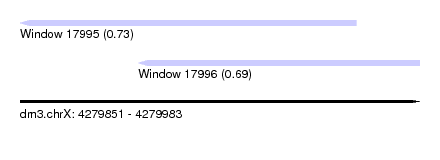
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,279,851 – 4,279,983 |
| Length | 132 |
| Max. P | 0.728867 |

| Location | 4,279,851 – 4,279,962 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Shannon entropy | 0.39416 |
| G+C content | 0.39609 |
| Mean single sequence MFE | -23.61 |
| Consensus MFE | -18.37 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4279851 111 - 22422827 GUGGUCUCGGGGAUCAUUCGCUUGGGGUCUCCUAACCAUCCGUACCUUAACAAAAAAAAAA-AA---AAAAAGUAAAAAUGUCGGCAUAUUUCUUGUUUUGGUUAUGCCACAUUU (((((...(((((((..........)))))))..)))))......................-..---.........((((((.((((((...((......)).)))))))))))) ( -24.70, z-score = -1.56, R) >droSim1.chrX 3299663 115 - 17042790 GUGGUCUCGGGGAUCAUUCGCUACGGGUCUCCCAACCAUCCGUACCUUAACAAAAAAAAAAAGAAGCAAAAACCAAAAAUGUCGGCAUAUUUCUUGUUUUGGUUAUGCCACAUUU (((((...(((((((..........)))))))..))))).....................................((((((.((((((...((......)).)))))))))))) ( -24.70, z-score = -0.80, R) >droSec1.super_4 3733564 112 + 6179234 GUGGUCUCGGGGAUCAUUCGCUUUGGGUCUCCUAACCAUCCGUACCUUAACAAAAAAAAGAUGA---AAAAAACAAAAAUGUCGGCAUAUUUCUGGUUUUGGUUAUGCCACAUUU (((((...(((((((..........)))))))..))))).........................---.........((((((.((((((...((......)).)))))))))))) ( -24.60, z-score = -0.70, R) >droYak2.chrX 17611661 97 + 21770863 GUGGUCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAACCUUAAC---AAAAAAG------GAAAACCAAAUAUGUCGGCAUAUUUC---------UUAUGCCACAUUU ((((.(((..((........))..)))...))))..........((((...---....)))------)..........((((.((((((....---------.)))))))))).. ( -23.30, z-score = -1.39, R) >droEre2.scaffold_4690 1655460 109 - 18748788 GUGGUCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAGCCUUGACAAAAAAAAAG------AAUAACCAAAAAUGUCGGCAUAUUUCUUGUUUCCAUUAUGCCACAUUU ((((((.....))))))..(((((((((......)))..))))))................------.........((((((.((((((..............)))))))))))) ( -25.24, z-score = -0.98, R) >droAna3.scaffold_13335 3146981 108 + 3335858 GUGGUCCUUUGGAACCUACGCUUGAGGUCUCCGUACCUCACAAUAAAAAACGUAAAAAAA-------AUAUGAAAAAACUGUCGGCAUAUUUCUUGUUUUCAUUAUGCCACAUUU (((((((...)).))).))...((((((......))))))..........((((......-------.)))).......(((.((((((..............)))))))))... ( -19.14, z-score = -1.03, R) >consensus GUGGUCUCGGGGAUCAUUCGCUUGGGGUCUCCAAACCAUCCGAACCUUAACAAAAAAAAAA______AAAAACCAAAAAUGUCGGCAUAUUUCUUGUUUUGGUUAUGCCACAUUU (((((...(((((((..........)))).))).))))).....................................((((((.((((((..............)))))))))))) (-18.37 = -18.46 + 0.09)
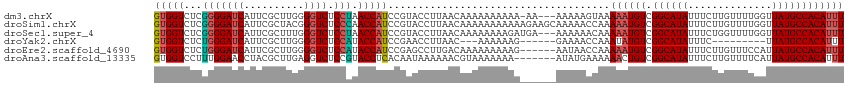
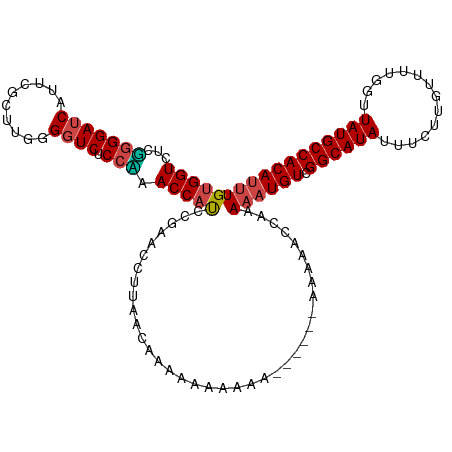
| Location | 4,279,890 – 4,279,983 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.70 |
| Shannon entropy | 0.52215 |
| G+C content | 0.45125 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -14.94 |
| Energy contribution | -16.27 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4279890 93 - 22422827 --------UAGGUCCUACAAACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUUGGGGUCUCCUAACCAUCCGUACCUUAACAAAAAAAAAAAAA---AAAAGUAA- --------(((((.......))))).((((((((...(((((((..........)))))))..)))).)))).....................---........- ( -22.40, z-score = -1.00, R) >droSim1.chrX 3299702 96 - 17042790 --------AAGGACCUAC-AACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUACGGGUCUCCCAACCAUCCGUACCUUAACAAAAAAAAAAAGAAGCAAAAACCAA --------((((.((((.-....))))(((((((...(((((((..........)))))))..)))).)))..))))............................ ( -22.30, z-score = -0.58, R) >droSec1.super_4 3733603 93 + 6179234 --------AAGGACCUAC-AACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUUUGGGUCUCCUAACCAUCCGUACCUUAACAAAAAAAAGAUGAA--AAAAACAA- --------((((.((((.-....))))(((((((...(((((((..........)))))))..)))).)))..)))).................--........- ( -22.30, z-score = -0.65, R) >droYak2.chrX 17611691 87 + 21770863 --------AGGGCCCUAA-AAACUAGGCGGUGGUCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAACCUUAACAAAAAAGGAAAACCAA--------- --------.((((((((.-......((((((((((.....))))).)))))))))))))..............((((.......))))........--------- ( -23.21, z-score = -0.16, R) >droEre2.scaffold_4690 1655499 90 - 18748788 --------AGGGUCCUAA-AAACUAGGCGGUGGUCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAGCCUUGACAAAAAAAAAGAAUAACCAA------ --------((((..(...-...(((((((((((((.....))))).))))))))(((......)))....)..))))......................------ ( -23.00, z-score = -0.25, R) >droAna3.scaffold_13335 3147020 98 + 3335858 AAAAAAAAGGGGCGGGGAAAACCUAGGCGGUGGUCCUUUGGAACCUACGCUUGAGGUCUCCGUACCUCACAAUAAAAAACGUAAAAAAAAUAUGAAAA------- .......(((..((((((...((((((((..(((((...)).)))..))))).)))))))))..)))............((((.......))))....------- ( -27.90, z-score = -2.29, R) >consensus ________AAGGACCUAA_AACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUUGGGGUCUCCAAACCAUCCGAACCUUAACAAAAAAAAAAAAAAA_AAA______ ..........((.........((((((((((((((.....)))))).)))))))).........))....................................... (-14.94 = -16.27 + 1.33)

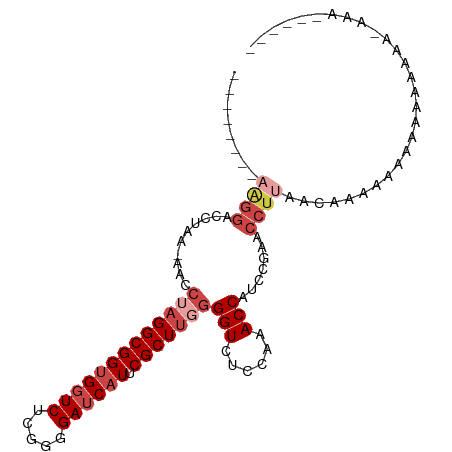
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:29 2011