
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,265,581 – 4,265,676 |
| Length | 95 |
| Max. P | 0.603265 |

| Location | 4,265,581 – 4,265,676 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Shannon entropy | 0.41332 |
| G+C content | 0.37825 |
| Mean single sequence MFE | -22.01 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.56 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
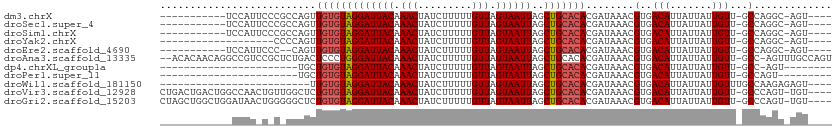
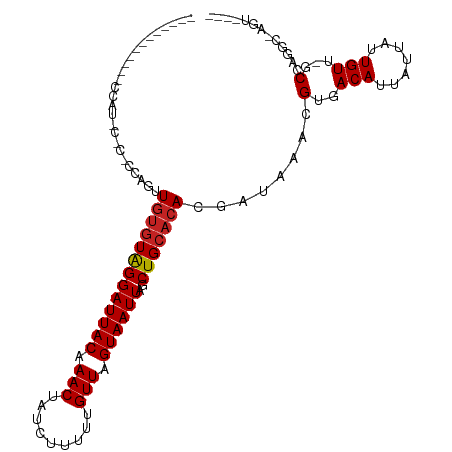
>dm3.chrX 4265581 95 + 22422827 -----------UCCAUUCCCGCCAGUUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCAGGC-AGU---- -----------.........(((.((.((((((((((((.(((.........))).))))))..)))))))).......(..(((.......))).-.)..)))-...---- ( -23.90, z-score = -2.20, R) >droSec1.super_4 3719822 95 - 6179234 -----------UCCAUUCCCGCCAGUUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCAGGC-AGU---- -----------.........(((.((.((((((((((((.(((.........))).))))))..)))))))).......(..(((.......))).-.)..)))-...---- ( -23.90, z-score = -2.20, R) >droSim1.chrX 3280263 95 + 17042790 -----------UCCAUUCCCGCCAGUUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCAGGC-AGU---- -----------.........(((.((.((((((((((((.(((.........))).))))))..)))))))).......(..(((.......))).-.)..)))-...---- ( -23.90, z-score = -2.20, R) >droYak2.chrX 17597789 87 - 21770863 -------------------CCCCAGUUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCAGGC-AGU---- -------------------..((.((.((((((((((((.(((.........))).))))))..)))))))).......(..(((.......))).-.)..)).-...---- ( -19.20, z-score = -0.99, R) >droEre2.scaffold_4690 1636043 93 + 18748788 -----------UCCAUUCCC--CAGUUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCAGGC-AGU---- -----------........(--(.((.((((((((((((.(((.........))).))))))..)))))))).......(..(((.......))).-.)..)).-...---- ( -19.20, z-score = -0.94, R) >droAna3.scaffold_13335 3130055 108 - 3335858 --ACACAACAGGCCGUCCGCUCUGACUCCCUGGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCC-AGUUUGCCAGU --......(((((.....)).))).....((((((((((.(((.........))).))))))(((((((.(((((((...........))))))))-).)-))))..)))). ( -22.10, z-score = -0.18, R) >dp4.chrXL_group1a 3796874 79 + 9151740 -----------------------UGCUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCC-AGU-------- -----------------------...(((((((((((((.(((.........))).))))))..)))))))........(..(((.......))).-.).-...-------- ( -17.70, z-score = -1.10, R) >droPer1.super_11 358770 79 + 2846995 -----------------------UGCUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCAGU--------- -----------------------...(((((((((((((.(((.........))).))))))..)))))))........(..(((.......))).-.)....--------- ( -17.70, z-score = -1.10, R) >droWil1.scaffold_181150 2858404 83 - 4952429 -------------------------UUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUUUGCCAAGAGAGU---- -------------------------.(((((((((((((.(((.........))).))))))..))))))).(.((((((............)))))).)........---- ( -15.00, z-score = -0.32, R) >droVir3.scaffold_12928 5274842 106 + 7717345 CUGACUGACUGGCCAACUGUUGGCUCUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCCAGU-UGU---- ......((((((......(((...(((((((((((((((.(((.........))).))))))..))))))).))..)))(..(((.......))).-.))))))-)..---- ( -28.00, z-score = -1.73, R) >droGri2.scaffold_15203 5745498 106 - 11997470 CUAGCUGGCUGGAUAACUGGGGGCUCUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU-GCCCAGU-UGU---- ((((....))))((((((((....(((((((((((((((.(((.........))).))))))..))))))).)).....(..(((.......))).-.))))))-)))---- ( -31.50, z-score = -2.18, R) >consensus ____________CCAU_C_C_CCAGUUGUGUAGGAUUACAAACUAUCUUUUUGUUAGUAAUUAGCUGCACACGAUAAACGUGACAUUAUUAUUGUU_GCCAGGC_AGU____ ...........................((((((((((((.(((.........))).))))))..))))))(((((((...........)))))))................. (-13.37 = -13.56 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:26 2011