| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,262,289 – 4,262,456 |
| Length | 167 |
| Max. P | 0.990117 |

| Location | 4,262,289 – 4,262,456 |
|---|---|
| Length | 167 |
| Sequences | 5 |
| Columns | 172 |
| Reading direction | reverse |
| Mean pairwise identity | 63.34 |
| Shannon entropy | 0.65082 |
| G+C content | 0.31610 |
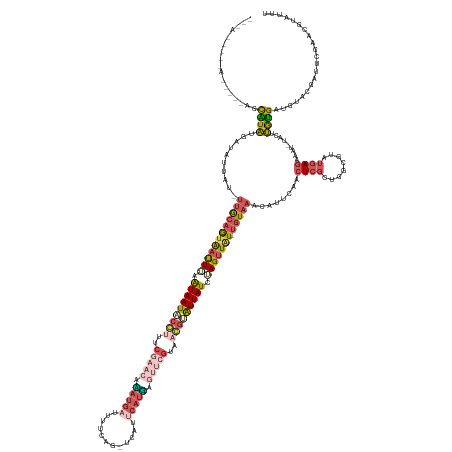
| Mean single sequence MFE | -37.76 |
| Consensus MFE | -15.32 |
| Energy contribution | -17.80 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
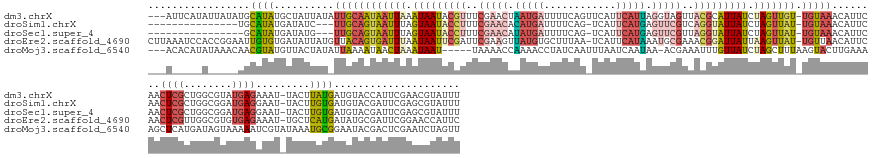
>dm3.chrX 4262289 167 - 22422827 ---AUUCAUAUUAUAUGCAUAUGCUAUUAUAUUGCAAUAAUUAAAUAAUACGUUUCGAACUAAUGAUUUUCAGUUCAUUCAUUAGGUAGUUACGCAUUAUCUAGUUGU-UGUAAACAUUCAACUCGCUGGCGUAUGAGAAAU-UACUUAUGAUGUACCAUUCGAACGUAUUU ---......(((((.((((((((....)))).))))))))).....((((((((.(((((((((((............)))))))((((((.(.(((...(((((.((-((........))))..)))))...))).).)))-))).............)))))))))))). ( -35.20, z-score = -1.55, R) >droSim1.chrX 3277296 151 - 17042790 ---------------UGCAUAUGAUAUC---UUGCAGUAAUUUAGUAAUACCUUUCGAACACAUGAUUUUCAG-UCAUUCAUGAGUUCGUCAGGUAUUAUCUAGUUAU-UGUAAACAUUCAACUCGCUGGCGGAUGAGGAAU-UACUUGUGAUGUACGAUUCGAGCGUAUUU ---------------......(((....---(((((((((((..(((((((((..(((((.(((((.......-....))))).)))))..)))))))))..))))))-)))))....)))...((((..(((((..(..((-((....))))...).)))))))))..... ( -44.00, z-score = -3.17, R) >droSec1.super_4 3716869 150 + 6179234 ----------------GCAUAUGAUAUG---UUGCAGUAAUUUAGUAAUACCUUUCGAACAUAUGAUUUUCAG-UCAUUCAUGAGUUCGUUAGGUAUUAUCUAGUUAU-UGUAAACAUUCAACUCGCUGGCGGAUGAGGAAU-UACUUGUGAUGUACGAUUCGAGCGUAUUU ----------------.....(((.(((---(((((((((((..(((((((((..(((((.(((((.......-....))))).)))))..)))))))))..))))))-))).))))))))...((((..(((((..(..((-((....))))...).)))))))))..... ( -44.70, z-score = -3.54, R) >droEre2.scaffold_4690 1633074 169 - 18748788 CUUAAAUCCACCGGAAUUGUGUGAUAUUAUGUUACAGUGAUUUAAUAAUUCGAUUCGAAGUUAUGUGCUUUAA-UCAUUCAUAAAUGCGAAACGGAUUAUUAAGUUAU-UGUUAACAUUCAACUCGUUGGCGUGUGAGAAAU-UGCUCAUGAUAUGCGAUUCGGAACCAUUC ..........(((.(((((((((.....((((((((((((((((((((((((.((((...(((((........-.....)))))...)))).))))))))))))))))-)).)))))).....((((.((((..........-)))).))))))))))))))))........ ( -47.52, z-score = -3.25, R) >droMoj3.scaffold_6540 31109847 163 - 34148556 ---ACACAUAUAAACAACGUAUGUUACUAUAUUAAAAUAACUAAAUAAU-----UAAAACCAAAACCUAUCAAUUUAAUCAAUAA-ACGAAAUUUGUUAUCUAGCUUUAAGUACUUGAAAAGCUCAUGAUAGUAAAAAUCGUAUAAAUGCGGAAUACGACUCGAAUCUAGUU ---..((((((.......))))))..............(((((......-----............((((((............(-((((...)))))....((((((..........))))))..))))))......((((((.........))))))........))))) ( -17.40, z-score = 0.40, R) >consensus ___A_____A_____AGCAUAUGAUAUUAU_UUGCAGUAAUUUAAUAAUACCUUUCGAACAAAUGAUUUUCAG_UCAUUCAUGAGUUCGUAACGUAUUAUCUAGUUAU_UGUAAACAUUCAACUCGCUGGCGUAUGAGAAAU_UACUUGUGAUGUACGAUUCGAACGUAUUU .................((((..........((((((((((((.(((((((((..(((((.(((((............))))).)))))..))))))))).))))))).)))))........((((........)))).........))))..................... (-15.32 = -17.80 + 2.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:25 2011