
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,260,155 – 4,260,356 |
| Length | 201 |
| Max. P | 0.896470 |

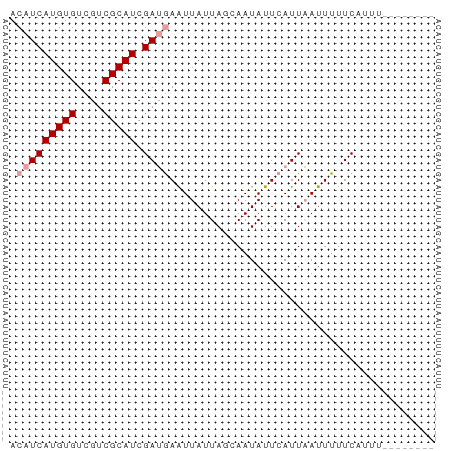
| Location | 4,260,155 – 4,260,212 |
|---|---|
| Length | 57 |
| Sequences | 3 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.26869 |
| G+C content | 0.30038 |
| Mean single sequence MFE | -7.36 |
| Consensus MFE | -6.07 |
| Energy contribution | -6.73 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
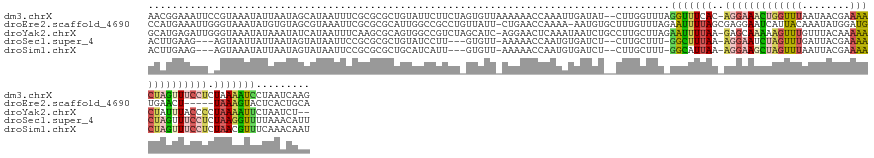
>dm3.chrX 4260155 57 - 22422827 ACAUCAUGUGUCGUCGCAUCGAGAAGUUAUUACCAAUAUUCAUUCAUUUUUCAUUUU------- .....(((((....))))).(((((((..................))))))).....------- ( -5.07, z-score = -0.48, R) >droSec1.super_4 3714777 52 + 6179234 ACAUCAUGUGUCGUCGCAUCGAUGAAUUAUUAGUAAUAUUCAUUAAUUUUUC------------ .(((((((((....))))).))))............................------------ ( -8.50, z-score = -1.53, R) >droSim1.chrX 3275195 64 - 17042790 ACAUCAUGUGUCGUCGCAUCGAUGAAUUAUUAGCAAUAUUCAUUAAUUUUUCAUUUCACUUUUC .(((((((((....))))).))))........................................ ( -8.50, z-score = -1.55, R) >consensus ACAUCAUGUGUCGUCGCAUCGAUGAAUUAUUAGCAAUAUUCAUUAAUUUUUCAUUU________ .(((((((((....))))).))))........................................ ( -6.07 = -6.73 + 0.67)


| Location | 4,260,212 – 4,260,356 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 147 |
| Reading direction | forward |
| Mean pairwise identity | 62.77 |
| Shannon entropy | 0.66022 |
| G+C content | 0.33622 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
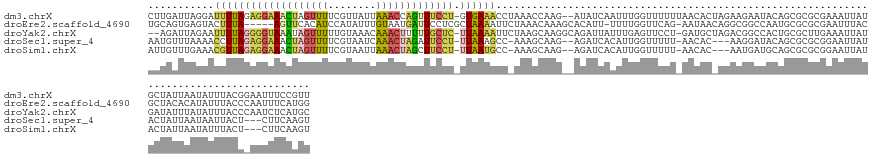
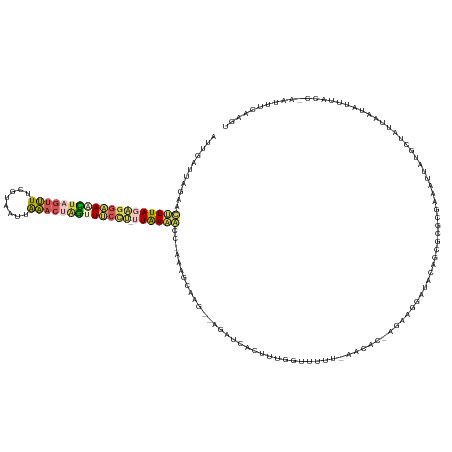
>dm3.chrX 4260212 144 + 22422827 CUUGAUUAGGAUUUUAGAGGAAACUAGUUUUCGUUAUUAAACCAGUUUCCU-GUGAAACCUAAACCAAG--AUAUCAAUUUGGUUUUUUAACACUAGAAGAAUACAGCGCGCGAAAUUAUGCUAUUAAUAUUUACGGAAUUUCCGUU ((((.(((((.(((((.((((((((.((((........)))).))))))))-.))))))))))..))))--........(((.(((((((....)))))))...))).(((.((((((.((.((........)))).))))))))). ( -33.70, z-score = -2.86, R) >droEre2.scaffold_4690 1630993 140 + 18748788 UGCAGUGAGUACUUUA-----AGUUCACAUCCAUAUUUGUAAUGAUUCCUCGCUAAAAUUCUAAACAAAGCACAUU-UUUUGGUUCAG-AAUAACAGGCGGCCAAUGCGCGCGAAUUUACGCUACACAUAUUUACCCAAUUUCAUGG ....((((((....((-----(((((..........((((....((((...(((((((..................-)))))))...)-))).))))(((((....)).)))))))))).))).)))........(((......))) ( -21.57, z-score = 1.01, R) >droYak2.chrX 17593044 143 - 21770863 --AGAUUAGAAUUUUAGGGGUAAAUAGUUUUUGUAAACAAACUUUUUGCUC-UUAAAAUUCUAAGCAAGGCAGAUUAUUUGAGUUCCU-GAUGCUAGACGGCCACUGCGCUUGAAAUUAUGAUAUUUAUAUUUACCCAAUCUCAUGC --...(((((((((((((((((((.(((((........))))).)))))))-)))))))))))).((((((((........(((....-...))).........)))).))))....(((((.(((...........))).))))). ( -34.43, z-score = -2.10, R) >droSec1.super_4 3714829 136 - 6179234 AAUGUUUAAAACCUUAGAGGAAACUAGUUUUCGUAAUCAAACUAGAUUCCU-UUAAAGCC-AAAGCAAG--AGAUCACAUUGGUUUUU-AACAC---AAGGAUACAGCGCGCGGAAUUAUACUAUUAAUAAUUACU---CUUCAAGU ..((((.(((((((((((((((.(((((((........))))))).)))))-)))).((.-...))...--..........)))))).-)))).---.................((((((.......))))))...---........ ( -25.80, z-score = -1.58, R) >droSim1.chrX 3275259 136 + 17042790 AUUGUUUGAAACGUUAGAGGAAACUAGUUUUCGUAAUUAAACUAGCUUCCU-UUAAUGCC-AAAGCAAG--AGAUCACAUUGGUUUUU-AACAC---AAUGAUGCAGCGCGCGGAAUUAUACUAUUAAUAUUUACU---CUUCAAGU .((((((....(((((((((((.(((((((........))))))).)))))-))))))..-.))))))(--((....(((((((....-.)).)---)))).(((.....))).....................))---)....... ( -32.90, z-score = -2.73, R) >consensus AUUGAUUAGAACUUUAGAGGAAACUAGUUUUCGUAAUUAAACUAGUUUCCU_UUAAAACC_AAAGCAAG__AGAUCACUUUGGUUUUU_AACAC_AGAAGGAUACAGCGCGCGAAAUUAUGCUAUUAAUAUUUACC_AAUUUCAAGU ...........(((((((((((((((((((........))))))))))))).))))))......................................................................................... (-10.70 = -11.14 + 0.44)
| Location | 4,260,212 – 4,260,356 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 147 |
| Reading direction | reverse |
| Mean pairwise identity | 62.77 |
| Shannon entropy | 0.66022 |
| G+C content | 0.33622 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -10.08 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
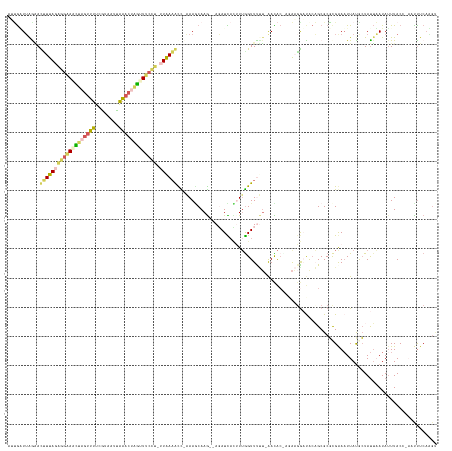
>dm3.chrX 4260212 144 - 22422827 AACGGAAAUUCCGUAAAUAUUAAUAGCAUAAUUUCGCGCGCUGUAUUCUUCUAGUGUUAAAAAACCAAAUUGAUAU--CUUGGUUUAGGUUUCAC-AGGAAACUGGUUUAAUAACGAAAACUAGUUUCCUCUAAAAUCCUAAUCAAG .((((.....))))..((((((((.((........))((((((........))))))...........))))))))--(((((((..(((((.(.-(((((((((((((........))))))))))))).).)))))..))))))) ( -38.90, z-score = -5.36, R) >droEre2.scaffold_4690 1630993 140 - 18748788 CCAUGAAAUUGGGUAAAUAUGUGUAGCGUAAAUUCGCGCGCAUUGGCCGCCUGUUAUU-CUGAACCAAAA-AAUGUGCUUUGUUUAGAAUUUUAGCGAGGAAUCAUUACAAAUAUGGAUGUGAACU-----UAAAGUACUCACUGCA (((((...((((((....((((((.(((......)))))))))..))).(((((((((-((((((.(((.-.......)))))))))))...)))).)))........))).)))))..((((((.-----....))..)))).... ( -32.10, z-score = -0.34, R) >droYak2.chrX 17593044 143 + 21770863 GCAUGAGAUUGGGUAAAUAUAAAUAUCAUAAUUUCAAGCGCAGUGGCCGUCUAGCAUC-AGGAACUCAAAUAAUCUGCCUUGCUUAGAAUUUUAA-GAGCAAAAAGUUUGUUUACAAAAACUAUUUACCCCUAAAAUUCUAAUCU-- ((.((((((((((((........)))).)))))))).))((((.(((.(..((.....-...........))..).)))))))(((((((((((.-(.(.(((.(((((........))))).))).).).)))))))))))...-- ( -29.59, z-score = -1.69, R) >droSec1.super_4 3714829 136 + 6179234 ACUUGAAG---AGUAAUUAUUAAUAGUAUAAUUCCGCGCGCUGUAUCCUU---GUGUU-AAAAACCAAUGUGAUCU--CUUGCUUU-GGCUUUAA-AGGAAUCUAGUUUGAUUACGAAAACUAGUUUCCUCUAAGGUUUUAAACAUU .......(---.(.((((((.......)))))).).).............---(((((-.((((((..........--...((...-.)).(((.-(((((.(((((((........))))))).))))).))))))))).))))). ( -26.10, z-score = -0.89, R) >droSim1.chrX 3275259 136 - 17042790 ACUUGAAG---AGUAAAUAUUAAUAGUAUAAUUCCGCGCGCUGCAUCAUU---GUGUU-AAAAACCAAUGUGAUCU--CUUGCUUU-GGCAUUAA-AGGAAGCUAGUUUAAUUACGAAAACUAGUUUCCUCUAACGUUUCAAACAAU ..((((((---.((.....................((.((..(((.((((---(.((.-....)))))))......--..)))..)-)))..((.-(((((((((((((........))))))))))))).)))).))))))..... ( -29.40, z-score = -1.93, R) >consensus ACAUGAAAUU_AGUAAAUAUUAAUAGCAUAAUUUCGCGCGCUGUAUCCUUCU_GUGUU_AAAAACCAAAAUGAUCU__CUUGCUUU_GGUUUUAA_AGGAAACUAGUUUAAUUACGAAAACUAGUUUCCUCUAAAGUUCUAAACAAU .......................................................................................((((((...(((((((((((((........)))))))))))))..))))))......... (-10.08 = -10.72 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:24 2011