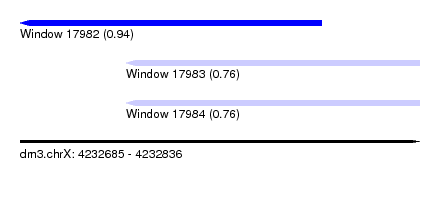
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,232,685 – 4,232,836 |
| Length | 151 |
| Max. P | 0.940355 |

| Location | 4,232,685 – 4,232,799 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.54136 |
| G+C content | 0.41602 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -17.71 |
| Energy contribution | -19.02 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4232685 114 - 22422827 GGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUUAGCCACUCCUUUAUCGUUGAUUUUGUGUGCUUCUCAUAUUAGCACUAUUAUCUAAGGCGAAACUAAUCUCC (((...((((((((((((((........))).))))))))))).))).........((((((((..((.(((((.........)))))))..)))...)))))........... ( -30.30, z-score = -1.71, R) >droEre2.scaffold_4690 1602799 114 - 18748788 GGCUGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCAGAUUUCAGCCACUCCCUAAUCGUUGAUUUUGUGUGCUUUUCAUAUUAGCACUAUUAUCUAAGGCGGCACUAAUCUCU ((((((((.....(((((((........)))))))....))))))))..............((((..(((((((((..(((.((....)).)))..))))).)))).))))... ( -32.90, z-score = -1.69, R) >droYak2.chrX 17562576 114 + 21770863 GGCUAAGAGGUCAGAGGGCACCUGCAUUUGCUCUUCUGAUUUUAGACACUUCUUUAUCGUUGAUUUUGUGUAUUUUUCAUAUAAACACUAUUAUCUAAGGCGGAACUAAUCUCU .....(((((((((((((((........))).)))))))).((((...((.(((((....((((..(((.(((.......))).)))..))))..))))).))..)))).)))) ( -22.10, z-score = -0.30, R) >droSec1.super_4 3684156 100 + 6179234 GGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGC--------------UGAUUCUGUGUGCCUUUCAUAUUGGCACUAUUAUCUAAGGCAAAACUAAUCUCC .(((..((((((((((((((........))).)))))))))))...--------------.(((.....(((((.........)))))....)))...)))............. ( -31.10, z-score = -2.28, R) >droSim1.chrX 3215922 100 - 17042790 GGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGC--------------UGAUUUUGUGUGCCUUUCAUAUUGGCACUAUUAUCUAAGGCAAAACUAAUCUCC .(((..((((((((((((((........))).)))))))))))...--------------.(((..((.(((((.........)))))))..)))...)))............. ( -31.80, z-score = -2.66, R) >droWil1.scaffold_181150 2816363 103 + 4952429 -GACACACGGCGAAUUUCCAUCUGCGAUUGGUUUUAAGUUUUUACC-------AUAUGCCUUAUACCACCCAACU-UCAGUUUAACAU--ACAUUAAAUGUGACGCGAACUUGC -........(((...........(((..((((...........)))-------)..)))................-(((((((((...--...)))))).))))))........ ( -10.80, z-score = 1.49, R) >consensus GGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGC_______UUAUCGUUGAUUUUGUGUGCCUUUCAUAUUAGCACUAUUAUCUAAGGCGAAACUAAUCUCC .(((..((((((((((((((........))).)))))))))))..................(((.....(((((.........)))))....)))...)))............. (-17.71 = -19.02 + 1.31)
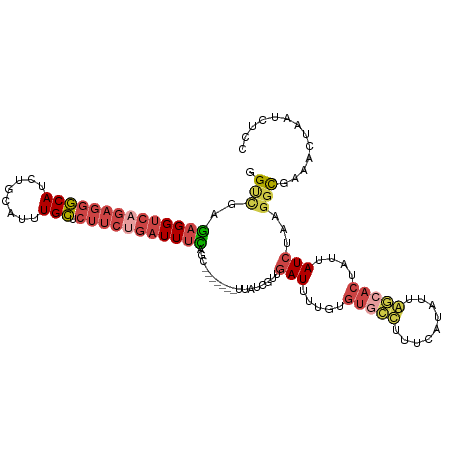
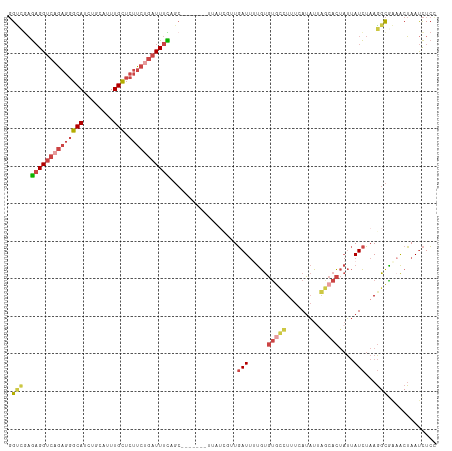
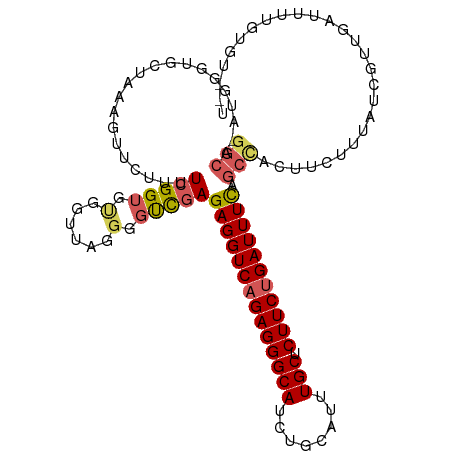
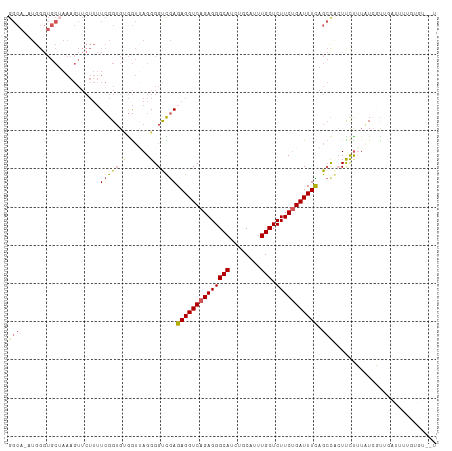
| Location | 4,232,725 – 4,232,836 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.35 |
| Shannon entropy | 0.50693 |
| G+C content | 0.47825 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -19.72 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4232725 111 - 22422827 GGCA-AUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUUAGCCACUCCUUUAUCGUUGAUUUUGUGUGCU ....-...((..((((((((.....(((((.....((((((.(.((((((((((((((........))).)))))))))))..).)))))).)))))..))))))).)..)) ( -33.60, z-score = -1.19, R) >droEre2.scaffold_4690 1602839 110 - 18748788 GGCA-AUGGGUGCAAAAGUUCUUUUCGGGG-GGUCAGGGGCUGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCAGAUUUCAGCCACUCCCUAAUCGUUGAUUUUGUGUGCU ....-..(.(..(((((.........((((-(((....((((((((.....(((((((........)))))))....))))))))))))))).........)))))..).). ( -37.47, z-score = -1.03, R) >droYak2.chrX 17562616 92 + 21770863 -------------------GGAAAUGGGUGCAA-AAGGGGCUAAGAGGUCAGAGGGCACCUGCAUUUGCUCUUCUGAUUUUAGACACUUCUUUAUCGUUGAUUUUGUGUAUU -------------------.(((((.((((..(-((((((....((((((((((((((........))).))))))))))).....)))))))..)))).)))))....... ( -22.80, z-score = -0.36, R) >droSec1.super_4 3684196 98 + 6179234 GGCAUUUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGCUGAUUCUGUGUGCC-------------- (((((.((((.(((...(..(((..(......)..)))..)...((((((((((((((........))).))))))))))))))....)))).)))))-------------- ( -31.20, z-score = -1.30, R) >droSim1.chrX 3215962 98 - 17042790 GGCAUUUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGCUGAUUUUGUGUGCC-------------- (((((((((.(.((((....(........)...)))).).))))((((((((((((((........))).)))))))))))............)))))-------------- ( -29.60, z-score = -1.07, R) >consensus GGCA_AUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGCCACUUCUUUAUCGUUGAUUUUGUGU__U (((.....................(((((.(......).)))))((((((((((((((........))).))))))))))).)))........................... (-19.72 = -20.12 + 0.40)

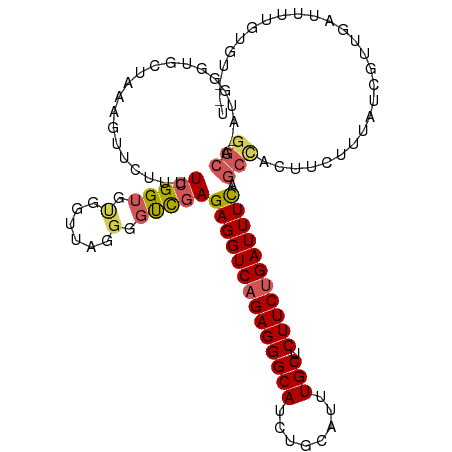
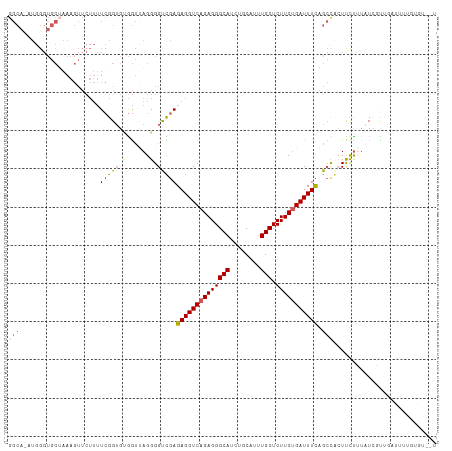
| Location | 4,232,725 – 4,232,836 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.35 |
| Shannon entropy | 0.50693 |
| G+C content | 0.47825 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -19.72 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4232725 111 - 22422827 GGCA-AUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUUAGCCACUCCUUUAUCGUUGAUUUUGUGUGCU ....-...((..((((((((.....(((((.....((((((.(.((((((((((((((........))).)))))))))))..).)))))).)))))..))))))).)..)) ( -33.60, z-score = -1.19, R) >droEre2.scaffold_4690 1602839 110 - 18748788 GGCA-AUGGGUGCAAAAGUUCUUUUCGGGG-GGUCAGGGGCUGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCAGAUUUCAGCCACUCCCUAAUCGUUGAUUUUGUGUGCU ....-..(.(..(((((.........((((-(((....((((((((.....(((((((........)))))))....))))))))))))))).........)))))..).). ( -37.47, z-score = -1.03, R) >droYak2.chrX 17562616 92 + 21770863 -------------------GGAAAUGGGUGCAA-AAGGGGCUAAGAGGUCAGAGGGCACCUGCAUUUGCUCUUCUGAUUUUAGACACUUCUUUAUCGUUGAUUUUGUGUAUU -------------------.(((((.((((..(-((((((....((((((((((((((........))).))))))))))).....)))))))..)))).)))))....... ( -22.80, z-score = -0.36, R) >droSec1.super_4 3684196 98 + 6179234 GGCAUUUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGCUGAUUCUGUGUGCC-------------- (((((.((((.(((...(..(((..(......)..)))..)...((((((((((((((........))).))))))))))))))....)))).)))))-------------- ( -31.20, z-score = -1.30, R) >droSim1.chrX 3215962 98 - 17042790 GGCAUUUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGCUGAUUUUGUGUGCC-------------- (((((((((.(.((((....(........)...)))).).))))((((((((((((((........))).)))))))))))............)))))-------------- ( -29.60, z-score = -1.07, R) >consensus GGCA_AUGGGUGCUAAAGUUCUUUUCGGUGUGGUUAGGGGUCGAGAGGUCAGAGGGCAUCUGCAUUUGCUCUUCUGAUUUCAGCCACUUCUUUAUCGUUGAUUUUGUGU__U (((.....................(((((.(......).)))))((((((((((((((........))).))))))))))).)))........................... (-19.72 = -20.12 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:19 2011